A Gene Regulatory Network for Cellular Reprogramming in Plant Regeneration
- PMID: 29462363
- PMCID: PMC6018650
- DOI: 10.1093/pcp/pcy013
A Gene Regulatory Network for Cellular Reprogramming in Plant Regeneration
Erratum in
-
Erratum.Plant Cell Physiol. 2018 Apr 1;59(4):876. doi: 10.1093/pcp/pcy075. Plant Cell Physiol. 2018. PMID: 29718476 Free PMC article. No abstract available.
Abstract
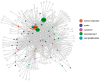
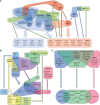
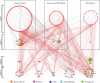
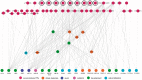
Wounding triggers organ regeneration in many plant species, and application of plant hormones, such as auxin and cytokinin, enhances their regenerative capacities in tissue culture. Recent studies have identified several key players mediating wound- and/or plant hormone-induced cellular reprogramming, but the global architecture of gene regulatory relationships underlying plant cellular reprogramming is still far from clear. In this study, we uncovered a gene regulatory network (GRN) associated with plant cellular reprogramming by using an enhanced yeast one-hybrid (eY1H) screen systematically to identify regulatory relationships between 252 transcription factors (TFs) and 48 promoters. Our network analyses suggest that wound- and/or hormone-invoked signals exhibit extensive cross-talk and regulate many common reprogramming-associated genes via multilayered regulatory cascades. Our data suggest that PLETHORA 3 (PLT3), ENHANCER OF SHOOT REGENERATION 1 (ESR1) and HEAT SHOCK FACTOR B 1 (HSFB1) act as critical nodes that have many overlapping targets and potentially connect upstream stimuli to downstream developmental decisions. Interestingly, a set of wound-inducible APETALA 2/ETHYLENE RESPONSE FACTORs (AP2/ERFs) appear to regulate these key genes, which, in turn, form feed-forward cascades that control downstream targets associated with callus formation and organ regeneration. In addition, we found another regulatory pathway, mediated by LATERAL ORGAN BOUNDARY/ASYMMETRIC LEAVES 2 (LOB/AS2) TFs, which probably plays a distinct but partially overlapping role alongside the AP2/ERFs in the putative gene regulatory cascades. Taken together, our findings provide the first global picture of the GRN governing plant cell reprogramming, which will serve as a valuable resource for future studies.
Figures

References
-
- Ahnert S.E. (2013) Power graph compression reveals dominant relationships in genetic transcription networks. Mol. Biosyst. 9: 2681–2685. - PubMed
-
- Assenov Y., Ramírez F., Schelhorn S.E., Lengauer T., Albrecht M. (2008) Computing topological parameters of biological networks. Bioinformatics 24: 282–284. - PubMed
-
- Atta R., Laurens L., Boucheron-Dubuisson E., Guivarc’h A., Carnero E., Giraudat-Pautot V., et al. (2009) Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J. 57: 626–644. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

