Truncated Glioma-Associated Oncogene Homolog 1 (tGLI1) Mediates Mesenchymal Glioblastoma via Transcriptional Activation of CD44
- PMID: 29463580
- PMCID: PMC5955849
- DOI: 10.1158/0008-5472.CAN-17-2933
Truncated Glioma-Associated Oncogene Homolog 1 (tGLI1) Mediates Mesenchymal Glioblastoma via Transcriptional Activation of CD44
Erratum in
-
Editor's Note: Truncated Glioma-Associated Oncogene Homolog 1 (tGLI1) Mediates Mesenchymal Glioblastoma via Transcriptional Activation of CD44.Cancer Res. 2024 Oct 1;84(19):3310. doi: 10.1158/0008-5472.CAN-24-2889. Cancer Res. 2024. PMID: 39350666 No abstract available.
Abstract
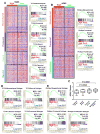
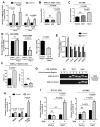
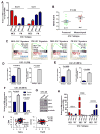
The molecular pathways driving mesenchymal glioblastoma (GBM) are still not well understood. We report here that truncated glioma-associated oncogene homolog 1 (tGLI1) is a tumor-specific transcription factor that facilitates GBM growth, is enriched in the mesenchymal subtype of GBM and glioma stem cells (GSC), and promotes mesenchymal GSC by upregulating transcription of CD44. In an orthotopic GBM xenograft mouse model, tGLI1-overexpressing tumors grew more aggressively with increased proliferation and angiogenesis compared with control and GLI1-overexpressing xenografts. tGLI1 was highly expressed in GBM clinical specimens but undetectable in normal brains, whereas GLI1 was expressed in both tissues. A tGLI1 activation signature (tGAS) correlated with glioma grade, tumor angiogenesis, and poor overall survival, and GBMs with high tGAS were enriched with mesenchymal GBM/GSC gene signatures. Neurospheres contained increased levels of tGLI1, but not GLI1, compared with the monolayer culture; mesenchymal GSC expressed more tGLI1 than proneural GSC. Ectopic tGLI1 expression enhanced the ability of mesenchymal GSC to yield neurospheres in vitro and to form tumors in mouse brains. Selective tGLI1 knockdown reduced neurosphere formation of GBM cells. tGLI1 bound to and transactivated the promoter of the CD44 gene, a marker and mediator for mesenchymal GSC, leading to its expression. Collectively, these findings advance our understanding of GBM biology by establishing tGLI1 as a novel transcriptional activator of CD44 and a novel mediator of mesenchymal GBM and GSC.Significance: These findings highlight the role of a tumor-specific gain-of-function transcription factor tGLI1 in mesenchymal glioma stem cell maintenance and mesenchymal GBM growth. Cancer Res; 78(10); 2589-600. ©2018 AACR.
©2018 American Association for Cancer Research.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–73. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

