Transcriptional Landscape and Regulatory Roles of Small Noncoding RNAs in the Oxidative Stress Response of the Haloarchaeon Haloferax volcanii
- PMID: 29463600
- PMCID: PMC5892119
- DOI: 10.1128/JB.00779-17
Transcriptional Landscape and Regulatory Roles of Small Noncoding RNAs in the Oxidative Stress Response of the Haloarchaeon Haloferax volcanii
Abstract
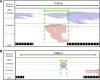
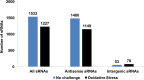
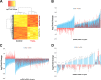
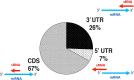
Haloarchaea in their natural environment are exposed to hypersalinity, intense solar radiation, and desiccation, all of which generate high levels of oxidative stress. Previous work has shown that haloarchaea are an order of magnitude more resistant to oxidative stress than most mesophilic organisms. Despite this resistance, the pathways haloarchaea use to respond to oxidative stress damage are similar to those of nonresistant organisms, suggesting that regulatory processes might be key to their robustness. Recently, small regulatory noncoding RNAs (sRNAs) were discovered in Archaea under a variety of environmental conditions. We report here the transcriptional landscape and functional roles of sRNAs in the regulation of the oxidative stress response of the model haloarchaeon Haloferax volcanii Thousands of sRNAs, both intergenic and antisense, were discovered using strand-specific sRNA sequencing (sRNA-seq), comprising 25 to 30% of the total transcriptome under no-challenge and oxidative stress conditions, respectively. We identified hundreds of differentially expressed sRNAs in response to hydrogen peroxide-induced oxidative stress in H. volcanii The targets of a group of antisense sRNAs decreased in expression when these sRNAs were upregulated, suggesting that sRNAs are potentially playing a negative regulatory role on mRNA targets at the transcript level. Target enrichment of these antisense sRNAs included mRNAs involved in transposon mobility, chemotaxis signaling, peptidase activity, and transcription factors.IMPORTANCE While a substantial body of experimental work has been done to uncover the functions of small regulatory noncoding RNAs (sRNAs) in gene regulation in Bacteria and Eukarya, the functional roles of sRNAs in Archaea are still poorly understood. This study is the first to establish the regulatory effects of sRNAs on mRNAs during the oxidative stress response in the haloarchaeon Haloferax volcanii Our work demonstrates that common principles for the response to a major cellular stress exist across the 3 domains of life while uncovering pathways that might be specific to the Archaea This work also underscores the relevance of sRNAs in adaptation to extreme environmental conditions.
Keywords: archaea; extreme environments; noncoding RNA; oxidative stress; small RNA; transcriptional regulation.
Copyright © 2018 Gelsinger and DiRuggiero.
Figures



References
-
- Robinson CK, Wierzchos J, Black C, Crits-Christoph A, Ma B, Ravel J, Ascaso C, Artieda O, Valea S, Roldan M, Gomez-Silva B, DiRuggiero J. 2015. Microbial diversity and the presence of algae in halite endolithic communities are correlated to atmospheric moisture in the hyper-arid zone of the Atacama Desert. Environ Microbiol 17:299–315. doi:10.1111/1462-2920.12364. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

