Mutant Isocitrate Dehydrogenase 1 Disrupts PKM2-β-Catenin-BRG1 Transcriptional Network-Driven CD47 Expression
- PMID: 29463646
- PMCID: PMC5902591
- DOI: 10.1128/MCB.00001-18
Mutant Isocitrate Dehydrogenase 1 Disrupts PKM2-β-Catenin-BRG1 Transcriptional Network-Driven CD47 Expression
Abstract
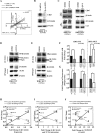
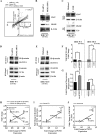
A gain-of-function mutation in isocitrate dehydrogenase 1 (IDH1) affects immune surveillance in gliomas. As elevated CD47 levels are associated with immune evasion in cancers, its status in gliomas harboring mutant IDH1 (IDH1-MT cells) was investigated. Decreased CD47 expression in IDH1-R132H-overexpressing cells was accompanied by diminished nuclear β-catenin, pyruvate kinase isoform M2 (PKM2), and TCF4 levels compared to those in cells harboring wild-type IDH1 (IDH1-WT cells). The inhibition of β-catenin in IDH1-WT cells abrogated CD47 expression, β-catenin-TCF4 interaction, and the transactivational activity of β-catenin/TCF4. The reverse effect was observed in IDH1-MT cells upon the pharmacological elevation of nuclear β-catenin levels. Genetic and pharmacological manipulation of nuclear PKM2 levels in IDH1-WT and IDH1-MT cells suggested that PKM2 is a positive regulator of the β-catenin-TCF4 interaction. The Cancer Genome Atlas (TCGA) data sets indicated diminished CD47, PKM2, and β-catenin levels in IDH1-MT gliomas compared to IDH1-WT gliomas. Also, elevated BRG1 levels with mutations in the ATP-dependent chromatin-remodeling site were observed in IDH1-MT glioma. The ectopic expression of ATPase-deficient BRG1 diminished CD47 expression as well as TCF4 occupancy on its promoter. Sequential chromatin immunoprecipitation (ChIP-re-ChIP) revealed the recruitment of the PKM2-β-catenin-BRG1-TCF4 complex to the TCF4 site on the CD47 promoter. This occupancy translated into CD47 transcription, as a diminished recruitment of this complex was observed in glioma cells bearing IDH1-R132H. In addition to its involvement in CD47 transcriptional regulation, PKM2-β-catenin-BRG1 cross talk affected the phagocytosis of IDH1-MT cells by microglia.
Keywords: BRG1; CD47; IDH1; PKM2; β-catenin.
Copyright © 2018 American Society for Microbiology.
Figures





References
-
- Bleeker FE, Atai NA, Lamba S, Jonker A, Rijkeboer D, Bosch KS, Tigchelaar W, Troost D, Vandertop WP, Bardelli A, Van Noorden CJ. 2010. The prognostic IDH1(R132) mutation is associated with reduced NADP+-dependent IDH activity in glioblastoma. Acta Neuropathol 119:487–494. doi:10.1007/s00401-010-0645-6. - DOI - PMC - PubMed
-
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. 2008. An integrated genomic analysis of human glioblastoma multiforme. Science 321:1807–1812. doi:10.1126/science.1164382. - DOI - PMC - PubMed
-
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. 2009. IDH1 and IDH2 mutations in gliomas. N Engl J Med 360:765–773. doi:10.1056/NEJMoa0808710. - DOI - PMC - PubMed
-
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, Thompson CB, Kaufman A, Guryanova O, Levine R, Heguy A, Viale A, Morris LG, Huse JT, Mellinghoff IK, Chan TA. 2012. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483:479–483. doi:10.1038/nature10866. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
