Ligand-Dependent Corepressor (LCoR) Is a Rexinoid-Inhibited Peroxisome Proliferator-Activated Receptor γ-Retinoid X Receptor α Coactivator
- PMID: 29463649
- PMCID: PMC5902596
- DOI: 10.1128/MCB.00107-17
Ligand-Dependent Corepressor (LCoR) Is a Rexinoid-Inhibited Peroxisome Proliferator-Activated Receptor γ-Retinoid X Receptor α Coactivator
Abstract
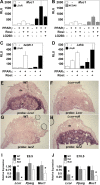
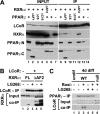
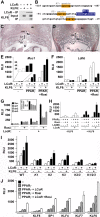
The nuclear receptor peroxisome proliferator-activated receptor gamma (PPARγ) is an essential regulator of placental development. To gain deeper insights into placental PPARγ signaling, we dissected its regulation of the Muc1 promoter. We find that, unlike prototypic target activation by heterodimeric receptors, which is either stimulated by or refractory to retinoid X receptor (RXR) ligands (rexinoids), the induction of Muc1 by liganded PPARγ requires RXRα but is inhibited by rexinoids. We demonstrate that this inhibition is mediated by the activation function 2 (AF2) domain of RXRα and that Muc1 activation entails altered AF2 structures of both PPARγ and RXRα. This unique regulation of Muc1 reflects specific coactivation of PPARγ-RXRα heterodimers by the transcription cofactor ligand-dependent corepressor (LCoR), corroborated by significant downregulation of Muc1 in Lcor-null placentas. LCoR interacts with PPARγ and RXRα in a synergistic fashion via adjacent noncanonical protein motifs, and the AF2 domain of ligand-bound RXRα inhibits this interaction. We further identify the transcription factor Krüppel-like factor 6 (KLF6) as a critical regulator of placental development and a component of Muc1 regulation in cooperation with PPARγ, RXRα, and LCoR. Combined, these studies reveal new principles and players in nuclear receptor function in general and placental PPARγ signaling in particular.
Keywords: KLF6; LCoR; PPARgamma; RXR; coactivators; rexinoids; transcription regulation.
Copyright © 2018 American Society for Microbiology.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
