Emergence of an Extensively Drug-Resistant Salmonella enterica Serovar Typhi Clone Harboring a Promiscuous Plasmid Encoding Resistance to Fluoroquinolones and Third-Generation Cephalosporins
- PMID: 29463654
- PMCID: PMC5821095
- DOI: 10.1128/mBio.00105-18
Emergence of an Extensively Drug-Resistant Salmonella enterica Serovar Typhi Clone Harboring a Promiscuous Plasmid Encoding Resistance to Fluoroquinolones and Third-Generation Cephalosporins
Abstract
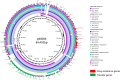
Antibiotic resistance is a major problem in Salmonella enterica serovar Typhi, the causative agent of typhoid. Multidrug-resistant (MDR) isolates are prevalent in parts of Asia and Africa and are often associated with the dominant H58 haplotype. Reduced susceptibility to fluoroquinolones is also widespread, and sporadic cases of resistance to third-generation cephalosporins or azithromycin have also been reported. Here, we report the first large-scale emergence and spread of a novel S Typhi clone harboring resistance to three first-line drugs (chloramphenicol, ampicillin, and trimethoprim-sulfamethoxazole) as well as fluoroquinolones and third-generation cephalosporins in Sindh, Pakistan, which we classify as extensively drug resistant (XDR). Over 300 XDR typhoid cases have emerged in Sindh, Pakistan, since November 2016. Additionally, a single case of travel-associated XDR typhoid has recently been identified in the United Kingdom. Whole-genome sequencing of over 80 of the XDR isolates revealed remarkable genetic clonality and sequence conservation, identified a large number of resistance determinants, and showed that these isolates were of haplotype H58. The XDR S Typhi clone encodes a chromosomally located resistance region and harbors a plasmid encoding additional resistance elements, including the blaCTX-M-15 extended-spectrum β-lactamase, and carrying the qnrS fluoroquinolone resistance gene. This antibiotic resistance-associated IncY plasmid exhibited high sequence identity to plasmids found in other enteric bacteria isolated from widely distributed geographic locations. This study highlights three concerning problems: the receding antibiotic arsenal for typhoid treatment, the ability of S Typhi to transform from MDR to XDR in a single step by acquisition of a plasmid, and the ability of XDR clones to spread globally.IMPORTANCE Typhoid fever is a severe disease caused by the Gram-negative bacterium Salmonella enterica serovar Typhi. Antibiotic-resistant S Typhi strains have become increasingly common. Here, we report the first large-scale emergence and spread of a novel extensively drug-resistant (XDR) S Typhi clone in Sindh, Pakistan. The XDR S Typhi is resistant to the majority of drugs available for the treatment of typhoid fever. This study highlights the evolving threat of antibiotic resistance in S Typhi and the value of antibiotic susceptibility testing and whole-genome sequencing in understanding emerging infectious diseases. We genetically characterized the XDR S Typhi to investigate the phylogenetic relationship between these isolates and a global collection of S Typhi isolates and to identify multiple genes linked to antibiotic resistance. This S Typhi clone harbored a promiscuous antibiotic resistance plasmid previously identified in other enteric bacteria. The increasing antibiotic resistance in S Typhi observed here adds urgency to the need for typhoid prevention measures.
Keywords: Salmonella; Typhi; antibiotic resistance; typhoid.
Copyright © 2018 Klemm et al.
Figures


Comment in
-
The Gathering Storm: Is Untreatable Typhoid Fever on the Way?mBio. 2018 Mar 20;9(2):e00482-18. doi: 10.1128/mBio.00482-18. mBio. 2018. PMID: 29559573 Free PMC article.
References
-
- Wong VK, Baker S, Pickard DJ, Parkhill J, Page AJ, Feasey NA, Kingsley RA, Thomson NR, Keane JA, Weill FX, Edwards DJ, Hawkey J, Harris SR, Mather AE, Cain AK, Hadfield J, Hart PJ, Thieu NT, Klemm EJ, Glinos DA, Breiman RF, Watson CH, Kariuki S, Gordon MA, Heyderman RS, Okoro C, Jacobs J, Lunguya O, Edmunds WJ, Msefula C, Chabalgoity JA, Kama M, Jenkins K, Dutta S, Marks F, Campos J, Thompson C, Obaro S, MacLennan CA, Dolecek C, Keddy KH, Smith AM, Parry CM, Karkey A, Mulholland EK, Campbell JI, Dongol S, Basnyat B, Dufour M, Bandaranayake D. 2015. Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella Typhi identifies inter- and intracontinental transmission events. Nat Genet 47:632–639. doi: 10.1038/ng.3281. - DOI - PMC - PubMed
-
- Feasey NA, Gaskell K, Wong V, Msefula C, Selemani G, Kumwenda S, Allain TJ, Mallewa J, Kennedy N, Bennett A, Nyirongo JO, Nyondo PA, Zulu MD, Parkhill J, Dougan G, Gordon MA, Heyderman RS. 2015. Rapid emergence of multidrug resistant, H58-lineage Salmonella typhi in Blantyre, Malawi. PLoS Negl Trop Dis 9:e0003748. doi: 10.1371/journal.pntd.0003748. - DOI - PMC - PubMed
-
- Hendriksen RS, Leekitcharoenphon P, Lukjancenko O, Lukwesa-Musyani C, Tambatamba B, Mwaba J, Kalonda A, Nakazwe R, Kwenda G, Jensen JD, Svendsen CA, Dittmann KK, Kaas RS, Cavaco LM, Aarestrup FM, Hasman H, Mwansa JC. 2015. Genomic signature of multidrug-resistant Salmonella enterica serovar typhi isolates related to a massive outbreak in Zambia between 2010 and 2012. J Clin Microbiol 53:262–272. doi: 10.1128/JCM.02026-14. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
