A set of regulatory genes co-expressed in embryonic human brain is implicated in disrupted speech development
- PMID: 29463886
- PMCID: PMC6756287
- DOI: 10.1038/s41380-018-0020-x
A set of regulatory genes co-expressed in embryonic human brain is implicated in disrupted speech development
Abstract
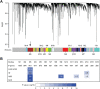
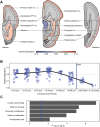
Genetic investigations of people with impaired development of spoken language provide windows into key aspects of human biology. Over 15 years after FOXP2 was identified, most speech and language impairments remain unexplained at the molecular level. We sequenced whole genomes of nineteen unrelated individuals diagnosed with childhood apraxia of speech, a rare disorder enriched for causative mutations of large effect. Where DNA was available from unaffected parents, we discovered de novo mutations, implicating genes, including CHD3, SETD1A and WDR5. In other probands, we identified novel loss-of-function variants affecting KAT6A, SETBP1, ZFHX4, TNRC6B and MKL2, regulatory genes with links to neurodevelopment. Several of the new candidates interact with each other or with known speech-related genes. Moreover, they show significant clustering within a single co-expression module of genes highly expressed during early human brain development. This study highlights gene regulatory pathways in the developing brain that may contribute to acquisition of proficient speech.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- American Speech-Language-Hearing Association. Childhood apraxia of speech. 2007. http://www.asha.org Accessed in April 2017.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

