Modifications and functional genomics of human transfer RNA
- PMID: 29463900
- PMCID: PMC5939049
- DOI: 10.1038/s41422-018-0013-y
Modifications and functional genomics of human transfer RNA
Abstract
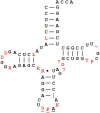
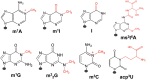
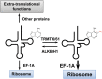
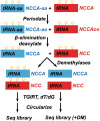
Transfer RNA (tRNA) is present at tens of millions of transcripts in a human cell and is the most abundant RNA in moles among all cellular RNAs. tRNA is also the most extensively modified RNA with, on an average, 13 modifications per molecule. The primary function of tRNA as the adaptor of amino acids and the genetic code in protein synthesis is well known. tRNA modifications play multi-faceted roles in decoding and other cellular processes. The abundance, modification, and aminoacylation (charging) levels of tRNAs contribute to mRNA decoding in ways that reflect the cell type and its environment; however, how these factors work together to maximize translation efficiency remains to be understood. tRNAs also interact with many proteins not involved in translation and this may coordinate translation activity and other processes in the cell. This review focuses on the modifications and the functional genomics of human tRNA and discusses future perspectives on the explorations of human tRNA biology.
Conflict of interest statement
The author declares that he has no conflict of interest.
Figures





References
-
- Gingold H, et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014;158:1281–1292. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

