Polyomavirus BK Nephropathy-Associated Transcriptomic Signatures: A Critical Reevaluation
- PMID: 29464200
- PMCID: PMC5811268
- DOI: 10.1097/TXD.0000000000000752
Polyomavirus BK Nephropathy-Associated Transcriptomic Signatures: A Critical Reevaluation
Abstract
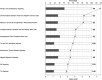
Background: Recent work using DNA microarrays has suggested that genes related to DNA replication, RNA polymerase assembly, and pathogen recognition receptors can serve as surrogate tissue biomarkers for polyomavirus BK nephropathy (BKPyVN).
Methods: We have examined this premise by looking for differential regulation of these genes using a different technology platform (RNA-seq) and an independent set 25 biopsies covering a wide spectrum of diagnoses.
Results: RNA-seq could discriminate T cell-mediated rejection from other common lesions seen in formalin fixed biopsy material. However, overlapping RNA-seq signatures were found among all disease processes investigated. Specifically, genes previously reported as being specific for the diagnosis of BKPyVN were found to be significantly upregulated in T cell-mediated rejection, inflamed areas of fibrosis/tubular atrophy, as well as acute tubular injury.
Conclusions: In conclusion, the search for virus specific molecular signatures is confounded by substantial overlap in pathogenetic mechanisms between BKPyVN and nonviral forms of allograft injury. Clinical heterogeneity, overlapping exposures, and different morphologic patterns and stage of disease are a source of substantial variability in "Omics" experiments. These variables should be better controlled in future biomarker studies on BKPyVN, T cell-mediated rejection, and other forms of allograft injury, before widespread implementation of these tests in the transplant clinic.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Hirsch HH, Randhawa P. BK polyomavirus in solid organ transplantation. Am J Transplant. 2013;13(Suppl4):179–188. - PubMed
-
- Randhawa P. Incorporation of pathology and laboratory findings into management algorithms for polyomavirus nephropathy. Am J Transplant. 2013;13:1379–1381. - PubMed
-
- Solez K, Axelsen RA, Benediktsson H, et al. International standardization of criteria for the histologic diagnosis of renal allograft rejection: the Banff working classification of kidney transplant pathology. Kidney Int. 1993;44:411–422. - PubMed
-
- Grinde B, Gayorfar M, Rinaldo CH. Impact of a polyomavirus (BKV) infection on mRNA expression in human endothelial cells. Virus Res. 2007;123:86–94. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

