The interplay of histone H2B ubiquitination with budding and fission yeast heterochromatin
- PMID: 29464330
- PMCID: PMC6039258
- DOI: 10.1007/s00294-018-0812-1
The interplay of histone H2B ubiquitination with budding and fission yeast heterochromatin
Abstract
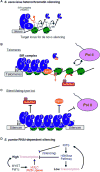
Mono-ubiquitinated histone H2B (H2B-Ub) is important for chromatin regulation of transcription, chromatin assembly, and also influences heterochromatin. In this review, we discuss the effects of H2B-Ub from nucleosome to higher-order chromatin structure. We then assess what is currently known of the role of H2B-Ub in heterochromatic silencing in budding and fission yeasts (S. cerevisiae and S. pombe), which have distinct silencing mechanisms. In budding yeast, the SIR complex initiates heterochromatin assembly with the aid of a H2B-Ub deubiquitinase, Ubp10. In fission yeast, the RNAi-dependent pathway initiates heterochromatin in the context of low H2B-Ub. We examine how the different silencing machineries overcome the challenge of H2B-Ub chromatin and highlight the importance of using these microorganisms to further our understanding of H2B-Ub in heterochromatic silencing pathways.
Keywords: Epigenetics; Heterochromatin; SIR complex; Silencing; Ubiquitination; Ubp10.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

