Single-Cell Droplet Microfluidic Screening for Antibodies Specifically Binding to Target Cells
- PMID: 29466744
- PMCID: PMC5842027
- DOI: 10.1016/j.celrep.2018.01.071
Single-Cell Droplet Microfluidic Screening for Antibodies Specifically Binding to Target Cells
Abstract
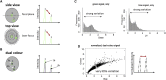
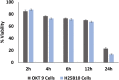
Monoclonal antibodies are a main player in modern drug discovery. Many antibody screening formats exist, each with specific advantages and limitations. Nonetheless, it remains challenging to screen antibodies for the binding of cell-surface receptors (the most important class of all drug targets) or for the binding to target cells rather than purified proteins. Here, we present a high-throughput droplet microfluidics approach employing dual-color normalized fluorescence readout to detect antibody binding. This enables us to obtain quantitative data on target cell recognition, using as little as 33 fg of IgG per assay. Starting with an excess of hybridoma cells releasing unspecific antibodies, individual clones secreting specific binders (of target cells co-encapsulated into droplets) could be enriched 220-fold after sorting 80,000 clones in a single experiment. This opens the way for therapeutic antibody discovery, especially since the single-cell approach is in principle also applicable to primary human plasma cells.
Keywords: FADS; antibody; antibody screening; droplet microfluidics; fluorescence activated droplet sorting; high-throughput; hybridoma; single-cell assay.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures






References
-
- Chaipan C., Pryszlak A., Dean H., Poignard P., Benes V., Griffiths A.D., Merten C.A. Single-virus droplet microfluidics for high-throughput screening of neutralizing epitopes on HIV particles. Cell Chem. Biol. 2017;24:751–757. - PubMed
-
- Chao G., Cochran J.R., Wittrup K.D. Fine epitope mapping of anti-epidermal growth factor receptor antibodies through random mutagenesis and yeast surface display. J. Mol. Biol. 2004;342:539–550. - PubMed
-
- Clausell-Tormos J., Lieber D., Baret J.C., El-Harrak A., Miller O.J., Frenz L., Blouwolff J., Humphry K.J., Köster S., Duan H. Droplet-based microfluidic platforms for the encapsulation and screening of Mammalian cells and multicellular organisms. Chem. Biol. 2008;15:427–437. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

