Massively Parallel Single Nucleus Transcriptional Profiling Defines Spinal Cord Neurons and Their Activity during Behavior
- PMID: 29466745
- PMCID: PMC5849084
- DOI: 10.1016/j.celrep.2018.02.003
Massively Parallel Single Nucleus Transcriptional Profiling Defines Spinal Cord Neurons and Their Activity during Behavior
Abstract
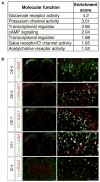
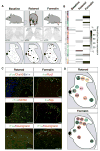
To understand the cellular basis of behavior, it is necessary to know the cell types that exist in the nervous system and their contributions to function. Spinal networks are essential for sensory processing and motor behavior and provide a powerful system for identifying the cellular correlates of behavior. Here, we used massively parallel single nucleus RNA sequencing (snRNA-seq) to create an atlas of the adult mouse lumbar spinal cord. We identified and molecularly characterized 43 neuronal populations. Next, we leveraged the snRNA-seq approach to provide unbiased identification of neuronal populations that were active following a sensory and a motor behavior, using a transcriptional signature of neuronal activity. This approach can be used in the future to link single nucleus gene expression data with dynamic biological responses to behavior, injury, and disease.
Keywords: neuronal activity; single cell RNA-seq; snRNA-seq; spinal cord.
Published by Elsevier Inc.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

