Transformation of Accessible Chromatin and 3D Nucleome Underlies Lineage Commitment of Early T Cells
- PMID: 29466755
- PMCID: PMC5847274
- DOI: 10.1016/j.immuni.2018.01.013
Transformation of Accessible Chromatin and 3D Nucleome Underlies Lineage Commitment of Early T Cells
Abstract
How chromatin reorganization coordinates differentiation and lineage commitment from hematopoietic stem and progenitor cells (HSPCs) to mature immune cells has not been well understood. Here, we carried out an integrative analysis of chromatin accessibility, topologically associating domains, AB compartments, and gene expression from HSPCs to CD4+CD8+ T cells. We found that abrupt genome-wide changes at all three levels of chromatin organization occur during the transition from double-negative stage 2 (DN2) to DN3, accompanying the T lineage commitment. The transcription factor BCL11B, a critical regulator of T cell commitment, is associated with increased chromatin interaction, and Bcl11b deletion compromised chromatin interaction at its target genes. We propose that these large-scale and concerted changes in chromatin organization present an energy barrier to prevent the cell from reversing its fate to earlier stages or redirecting to alternatives and thus lock the cell fate into the T lineages.
Keywords: 4D nucleome; AB compartment conversion; AD connectivity; BCL11B; DNase hypersensitive sites; T cell development; chromatin conformation; lineage commitment.
Published by Elsevier Inc.
Conflict of interest statement
The authors declare no competing interests.
Figures


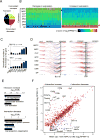
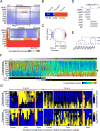

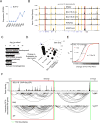

Comment in
-
T Cell LEGO: Identifying the Master Builders and What They Do.Immunity. 2018 Feb 20;48(2):185-187. doi: 10.1016/j.immuni.2018.02.004. Immunity. 2018. PMID: 29466746
References
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Bhandoola A, Sambandam A. From stem cell to T cell: one route or many? Nat Rev Immunol. 2006;6:117–126. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

