Yeast Aim21/Tda2 both regulates free actin by reducing barbed end assembly and forms a complex with Cap1/Cap2 to balance actin assembly between patches and cables
- PMID: 29467252
- PMCID: PMC5896931
- DOI: 10.1091/mbc.E17-10-0592
Yeast Aim21/Tda2 both regulates free actin by reducing barbed end assembly and forms a complex with Cap1/Cap2 to balance actin assembly between patches and cables
Abstract
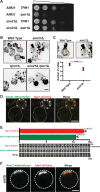
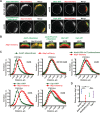
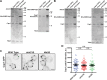
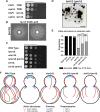
Yeast Aim21 is recruited by the SH3-containing proteins Bbc1 and Abp1 to patches and, with Tda2, reduces barbed end assembly to balance the distribution of actin between patches and cables. Aim21/Tda2 also interacts with Cap1/Cap2, revealing a complex interplay between actin assembly regulators.
Figures




References
-
- Amatruda JF, Cannon JF, Tatchell K, Hug C, Cooper JA. (1990). Disruption of the actin cytoskeleton in yeast capping protein mutants. Nature , 352–354. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

