High-throughput metabolomic analysis predicts mode of action of uncharacterized antimicrobial compounds
- PMID: 29467300
- PMCID: PMC6544516
- DOI: 10.1126/scitranslmed.aal3973
High-throughput metabolomic analysis predicts mode of action of uncharacterized antimicrobial compounds
Abstract
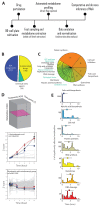
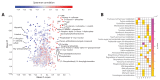
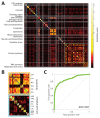
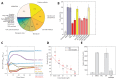
Rapidly spreading antibiotic resistance and the low discovery rate of new antimicrobial compounds demand more effective strategies for early drug discovery. One bottleneck in the drug discovery pipeline is the identification of the modes of action (MoAs) of new compounds. We have developed a rapid systematic metabolome profiling strategy to classify the MoAs of bioactive compounds. The method predicted MoA-specific metabolic responses in the nonpathogenic bacterium Mycobacterium smegmatis after treatment with 62 reference compounds with known MoAs and different metabolic and nonmetabolic targets. We then analyzed a library of 212 new antimycobacterial compounds with unknown MoAs from a drug discovery effort by the pharmaceutical company GlaxoSmithKline (GSK). More than 70% of these new compounds induced metabolic responses in M. smegmatis indicative of known MoAs, seven of which were experimentally validated. Only 8% (16) of the compounds appeared to target unconventional cellular processes, illustrating the difficulty in discovering new antibiotics with different MoAs among compounds used as monotherapies. For six of the GSK compounds with potentially new MoAs, the metabolome profiles suggested their ability to interfere with trehalose and lipid metabolism. This was supported by whole-genome sequencing of spontaneous drug-resistant mutants of the pathogen Mycobacterium tuberculosis and in vitro compound-proteome interaction analysis for one of these compounds. Our compendium of drug-metabolome profiles can be used to rapidly query the MoAs of uncharacterized antimicrobial compounds and should be a useful resource for the drug discovery community.
Copyright © 2018 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures

References
-
- Burdine L, Kodadek T. Target Identification in Chemical Genetics: The (Often) Missing Link. Chem Biol. 2004;11:593–597. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

