Universal Vaccines and Vaccine Platforms to Protect against Influenza Viruses in Humans and Agriculture
- PMID: 29467737
- PMCID: PMC5808216
- DOI: 10.3389/fmicb.2018.00123
Universal Vaccines and Vaccine Platforms to Protect against Influenza Viruses in Humans and Agriculture
Abstract
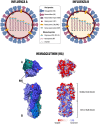
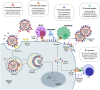
Influenza virus infections pose a significant threat to public health due to annual seasonal epidemics and occasional pandemics. Influenza is also associated with significant economic losses in animal production. The most effective way to prevent influenza infections is through vaccination. Current vaccine programs rely heavily on the vaccine's ability to stimulate neutralizing antibody responses to the hemagglutinin (HA) protein. One of the biggest challenges to an effective vaccination program lies on the fact that influenza viruses are ever-changing, leading to antigenic drift that results in escape from earlier immune responses. Efforts toward overcoming these challenges aim at improving the strength and/or breadth of the immune response. Novel vaccine technologies, the so-called universal vaccines, focus on stimulating better cross-protection against many or all influenza strains. However, vaccine platforms or manufacturing technologies being tested to improve vaccine efficacy are heterogeneous between different species and/or either tailored for epidemic or pandemic influenza. Here, we discuss current vaccines to protect humans and animals against influenza, highlighting challenges faced to effective and uniform novel vaccination strategies and approaches.
Keywords: immune response; influenza vaccines; live attenuated vaccines; poultry; swine; universal vaccines; vaccine platform; vectored vaccine.
Figures

References
-
- Anderson T. K., Macken C. A., Lewis N. S., Scheuermann R. H., Van Reeth K., Brown I. H., et al. (2016). A Phylogeny-Based Global Nomenclature System and Automated Annotation Tool for H1 Hemagglutinin Genes from Swine Influenza A Viruses. mSphere 1, e00275–e00216. 10.1128/mSphere.00275-16 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

