Fast phylogenetic inference from typing data
- PMID: 29467814
- PMCID: PMC5815242
- DOI: 10.1186/s13015-017-0119-7
Fast phylogenetic inference from typing data
Abstract
Background: Microbial typing methods are commonly used to study the relatedness of bacterial strains. Sequence-based typing methods are a gold standard for epidemiological surveillance due to the inherent portability of sequence and allelic profile data, fast analysis times and their capacity to create common nomenclatures for strains or clones. This led to development of several novel methods and several databases being made available for many microbial species. With the mainstream use of High Throughput Sequencing, the amount of data being accumulated in these databases is huge, storing thousands of different profiles. On the other hand, computing genetic evolutionary distances among a set of typing profiles or taxa dominates the running time of many phylogenetic inference methods. It is important also to note that most of genetic evolution distance definitions rely, even if indirectly, on computing the pairwise Hamming distance among sequences or profiles.
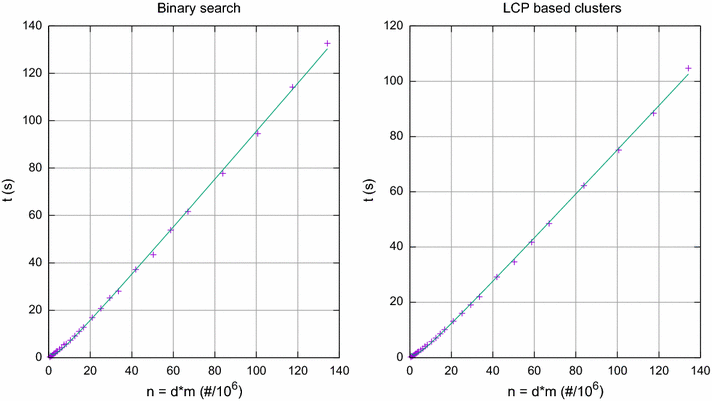
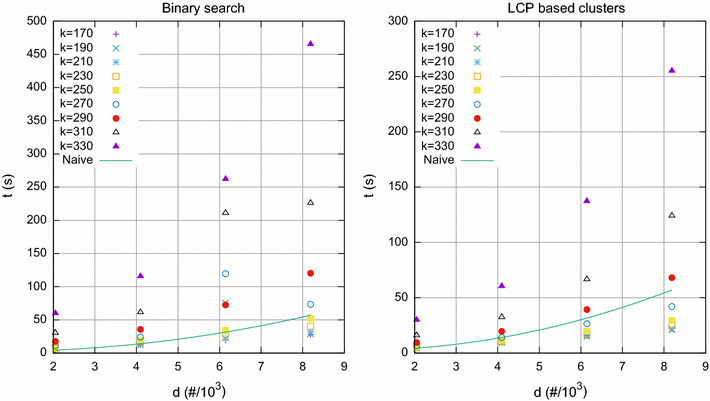
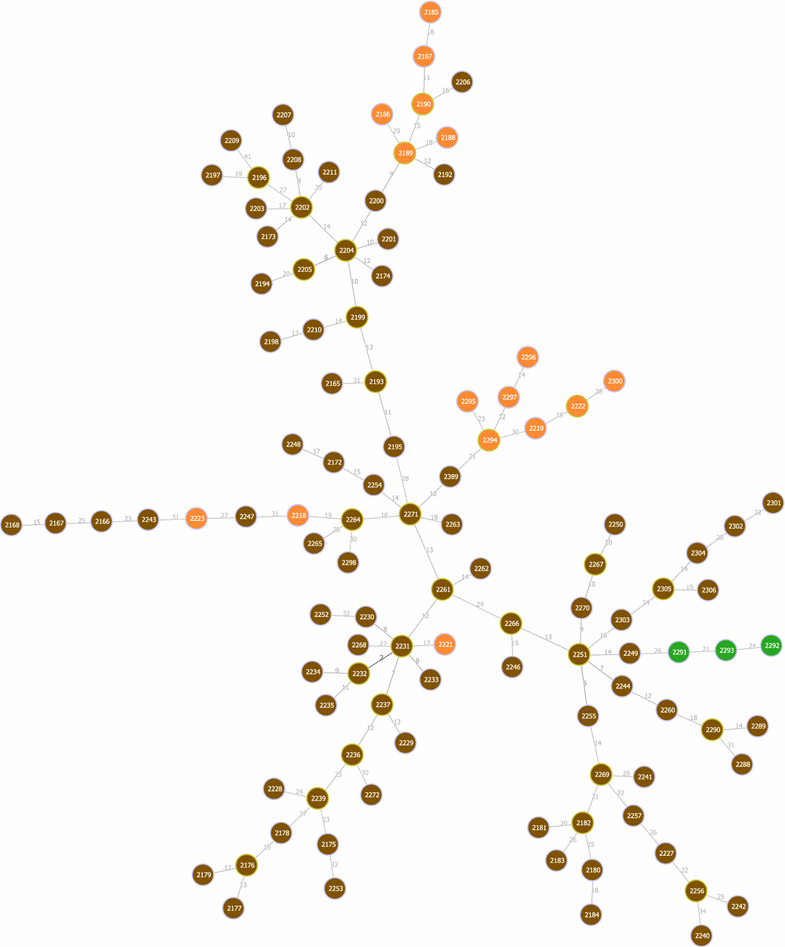
Results: We propose here an average-case linear-time algorithm to compute pairwise Hamming distances among a set of taxa under a given Hamming distance threshold. This article includes both a theoretical analysis and extensive experimental results concerning the proposed algorithm. We further show how this algorithm can be successfully integrated into a well known phylogenetic inference method, and how it can be used to speedup querying local phylogenetic patterns over large typing databases.
Keywords: Computational biology; Hamming distance; Phylogenetic inference.
Figures

References
-
- Maiden MC, Bygraves JA, Feil EJ, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95(6):3140–3145. doi: 10.1073/pnas.95.6.3140. - DOI - PMC - PubMed
-
- Huson DH, Rupp R, Scornavacca C. Phylogenetic networks: concepts, algorithms and applications. New York: Cambridge University Press; 2010.
-
- Robinson DA, Feil EJ. Bacterial population genetics in infectious disease. Hoboken: Wiley; 2010.
-
- Saitou N. Introduction to evolutionary genomics. London: Springer; 2013.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

