Synechocystis PCC 6803 overexpressing RuBisCO grow faster with increased photosynthesis
- PMID: 29468130
- PMCID: PMC5779733
- DOI: 10.1016/j.meteno.2017.02.002
Synechocystis PCC 6803 overexpressing RuBisCO grow faster with increased photosynthesis
Abstract
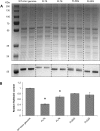
The ribulose-1,5-bisphosphate (RuBP) oxygenation reaction catalyzed by Ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) is competing with carboxylation, being negative for both energy and carbon balances in photoautotrophic organisms. This makes RuBisCO one of the bottlenecks for oxygenic photosynthesis and carbon fixation. In this study, RuBisCO was overexpressed in the unicellular cyanobacterium Synechocystis PCC 6803. Relative RuBisCO levels in the engineered strains FL50 and FL52 increased 2.1 times and 1.4 times, respectively, and both strains showed increased growth, photosynthesis and in vitro RuBisCO activity. The oxygen evolution rate increased by 54% and 42% on per chlorophyll basis, while the in vitro RuBisCO activity increased by 52% and 8.6%, respectively. The overexpressed RuBisCO were tagged with a FLAG tag, in strain FL50 on the N terminus of the large subunit while in strain FL52 on the C terminus of the small subunit. The presence of a FLAG tag enhanced transcription of the genes encoding RuBisCO, and, with high possibility, also enhanced the initiation of translation or stability of the enzyme. However, when using a streptavidin-binding tag II (strep-tag II), we did not observe a similar effect. Tagged RuBisCO offers an opportunity for further studying RuBisCO expression and stability. Increased levels of RuBisCO can further improve photosynthesis and growth in the cyanobacterium Synechocystis PCC 6803 under certain growth conditions.
Keywords: FLAG tag; Increased growth; Photosynthesis; RuBisCO; Strep-tag II; Synechocystis PCC 6803.
Figures






Similar articles
-
Effects of overexpressing photosynthetic carbon flux control enzymes in the cyanobacterium Synechocystis PCC 6803.Metab Eng. 2016 Nov;38:56-64. doi: 10.1016/j.ymben.2016.06.005. Epub 2016 Jun 18. Metab Eng. 2016. PMID: 27328433
-
The transcriptional regulator RbcR controls ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) genes in the cyanobacterium Synechocystis sp. PCC 6803.New Phytol. 2022 Jul;235(2):432-445. doi: 10.1111/nph.18139. Epub 2022 Apr 22. New Phytol. 2022. PMID: 35377491
-
Rubisco mutagenesis provides new insight into limitations on photosynthesis and growth in Synechocystis PCC6803.J Exp Bot. 2011 Aug;62(12):4173-82. doi: 10.1093/jxb/err116. Epub 2011 May 6. J Exp Bot. 2011. PMID: 21551078 Free PMC article.
-
RubisCO Early Oxygenase Activity: A Kinetic and Evolutionary Perspective.Bioessays. 2017 Nov;39(11). doi: 10.1002/bies.201700071. Epub 2017 Oct 4. Bioessays. 2017. PMID: 28976010 Review.
-
Biogenesis and Metabolic Maintenance of Rubisco.Annu Rev Plant Biol. 2017 Apr 28;68:29-60. doi: 10.1146/annurev-arplant-043015-111633. Epub 2017 Jan 11. Annu Rev Plant Biol. 2017. PMID: 28125284 Review.
Cited by
-
Advances in Genetic Engineering in Improving Photosynthesis and Microalgal Productivity.Int J Mol Sci. 2023 Jan 18;24(3):1898. doi: 10.3390/ijms24031898. Int J Mol Sci. 2023. PMID: 36768215 Free PMC article. Review.
-
Kinetic modeling of the Calvin cycle identifies flux control and stable metabolomes in Synechocystis carbon fixation.J Exp Bot. 2019 Feb 5;70(3):973-983. doi: 10.1093/jxb/ery382. J Exp Bot. 2019. PMID: 30371804 Free PMC article.
-
Photoautotrophic Growth Rate Enhancement of Synechocystis sp. PCC6803 by Heterologous Production of 2-Oxoglutarate:Ferredoxin Oxidoreductase from Chlorobaculum tepidum.Biology (Basel). 2022 Dec 29;12(1):59. doi: 10.3390/biology12010059. Biology (Basel). 2022. PMID: 36671751 Free PMC article.
-
Exploring the Role of a Cytokinin-Activating Enzyme LONELY GUY in Unicellular Microalga Chlorella variabilis.Front Plant Sci. 2021 Jan 29;11:611871. doi: 10.3389/fpls.2020.611871. eCollection 2020. Front Plant Sci. 2021. PMID: 33613586 Free PMC article.
-
Synthetic biology for space exploration.NPJ Microgravity. 2025 Jul 12;11(1):41. doi: 10.1038/s41526-025-00488-7. NPJ Microgravity. 2025. PMID: 40651964 Free PMC article. Review.
References
-
- Atsumi S., Higashide W., Liao J.C. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat. Biotechnol. 2009;27:1177–1180. - PubMed
-
- Ayala J.C., Pimienta E., Rodríguez C., Anne J., Vallin C., Milanes M.T., King-Batsios E., Huygen K., Van Mellaert L. Use of Strep-tag II for rapid detection and purification of Mycobacterium tuberculosis recombinant antigens secreted by Streptomyces lividans. J. Microbiol. Methods. 2013;94:192–198. - PubMed
-
- Carmo-Silva E., Scales J.C., Madgwick P.J., Parry M.A.J. Optimizing rubisco and its regulation for greater resource use efficiency. Plant Cell Environ. 2015;38:1817–1832. - PubMed
-
- Durall C., Lindblad P. Mechanisms of carbon fixation and engineering for increased carbon fixation in cyanobacteria. Algal Res. 2015;11:263–270.
LinkOut - more resources
Full Text Sources
Other Literature Sources

