Gene expression and ultrastructure of meso- and thermophilic methanotrophic consortia
- PMID: 29468803
- PMCID: PMC5947290
- DOI: 10.1111/1462-2920.14077
Gene expression and ultrastructure of meso- and thermophilic methanotrophic consortia
Abstract
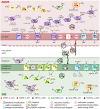
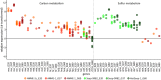
The sulfate-dependent, anaerobic oxidation of methane (AOM) is an important sink for methane in marine environments. It is carried out between anaerobic methanotrophic archaea (ANME) and sulfate-reducing bacteria (SRB) living in syntrophic partnership. In this study, we compared the genomes, gene expression patterns and ultrastructures of three phylogenetically different microbial consortia found in hydrocarbon-rich environments under different temperature regimes: ANME-1a/HotSeep-1 (60°C), ANME-1a/Seep-SRB2 (37°C) and ANME-2c/Seep-SRB2 (20°C). All three ANME encode a reverse methanogenesis pathway: ANME-2c encodes all enzymes, while ANME-1a lacks the gene for N5,N10-methylene tetrahydromethanopterin reductase (mer) and encodes a methylenetetrahydrofolate reductase (Met). The bacterial partners contain the genes encoding the canonical dissimilatory sulfate reduction pathway. During AOM, all three consortia types highly expressed genes encoding for the formation of flagella or type IV pili and/or c-type cytochromes, some predicted to be extracellular. ANME-2c expressed potentially extracellular cytochromes with up to 32 hemes, whereas ANME-1a and SRB expressed less complex cytochromes (≤ 8 and ≤ 12 heme respectively). The intercellular space of all consortia showed nanowire-like structures and heme-rich areas. These features are proposed to enable interspecies electron exchange, hence suggesting that direct electron transfer is a common mechanism to sulfate-dependent AOM, and that both partners synthesize molecules to enable it.
© 2018 The Authors. Environmental Microbiology published by Society for Applied Microbiology and John Wiley & Sons Ltd.
Figures


References
-
- Adhikari, R.Y. , Malvankar, N.S. , Tuominen, M.T. , and Lovley, D.R. (2015) Conductivity of individual Geobacter pili. RSC Adv 6: 8354–8357.
-
- Boetius, A. , and Wenzhöfer, F. (2013) Seafloor oxygen consumption fuelled by methane from cold seeps. Nat Geosci 6: 725–734.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

