Fragment-Based Discovery of Inhibitors of the Bacterial DnaG-SSB Interaction
- PMID: 29470422
- PMCID: PMC5872125
- DOI: 10.3390/antibiotics7010014
Fragment-Based Discovery of Inhibitors of the Bacterial DnaG-SSB Interaction
Abstract
In bacteria, the DnaG primase is responsible for synthesis of short RNA primers used to initiate chain extension by replicative DNA polymerase(s) during chromosomal replication. Among the proteins with which Escherichia coli DnaG interacts is the single-stranded DNA-binding protein, SSB. The C-terminal hexapeptide motif of SSB (DDDIPF; SSB-Ct) is highly conserved and is known to engage in essential interactions with many proteins in nucleic acid metabolism, including primase. Here, fragment-based screening by saturation-transfer difference nuclear magnetic resonance (STD-NMR) and surface plasmon resonance assays identified inhibitors of the primase/SSB-Ct interaction. Hits were shown to bind to the SSB-Ct-binding site using 15N-¹H HSQC spectra. STD-NMR was used to demonstrate binding of one hit to other SSB-Ct binding partners, confirming the possibility of simultaneous inhibition of multiple protein/SSB interactions. The fragment molecules represent promising scaffolds on which to build to discover new antibacterial compounds.
Keywords: SSB; antibacterial agents; fragment-based screening; primase; protein–protein interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

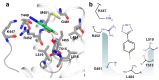




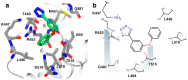

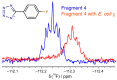
References
-
- Kobori J.A., Kornberg A. The Escherichia coli dnaC gene product. II. Purification, physical properties, and role in replication. J. Biol. Chem. 1982;257:13763–13769. - PubMed
-
- Kobori J.A., Kornberg A. The Escherichia coli dnaC gene product. III. Properties of the dnaB–dnaC protein complex. J. Biol. Chem. 1982;257:13770–13775. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

