Most yeast SH3 domains bind peptide targets with high intrinsic specificity
- PMID: 29470497
- PMCID: PMC5823434
- DOI: 10.1371/journal.pone.0193128
Most yeast SH3 domains bind peptide targets with high intrinsic specificity
Abstract
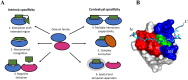
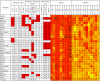
A need exists to develop bioinformatics for predicting differences in protein function, especially for members of a domain family who share a common fold, yet are found in a diverse array of proteins. Many domain families have been conserved over large evolutionary spans and representative genomic data during these periods are now available. This allows a simple method for grouping domain sequences to reveal common and unique/specific binding residues. As such, we hypothesize that sequence alignment analysis of the yeast SH3 domain family across ancestral species in the fungal kingdom can determine whether each member encodes specific information to bind unique peptide targets. With this approach, we identify important specific residues for a given domain as those that show little conservation within an alignment of yeast domain family members (paralogs) but are conserved in an alignment of its direct relatives (orthologs). We find most of the yeast SH3 domain family members have maintained unique amino acid conservation patterns that suggest they bind peptide targets with high intrinsic specificity through varying degrees of non-canonical recognition. For a minority of domains, we predict a less diverse binding surface, likely requiring additional factors to bind targets specifically. We observe that our predictions are consistent with high throughput binding data, which suggests our approach can probe intrinsic binding specificity in any other interaction domain family that is maintained during evolution.
Conflict of interest statement
Figures




Similar articles
-
Evolution of the SH3 Domain Specificity Landscape in Yeasts.PLoS One. 2015 Jun 11;10(6):e0129229. doi: 10.1371/journal.pone.0129229. eCollection 2015. PLoS One. 2015. PMID: 26068101 Free PMC article.
-
Recognition of non-canonical peptides by the yeast Fus1p SH3 domain: elucidation of a common mechanism for diverse SH3 domain specificities.J Mol Biol. 2008 Mar 28;377(3):889-901. doi: 10.1016/j.jmb.2008.01.063. Epub 2008 Jan 31. J Mol Biol. 2008. PMID: 18280496
-
Structure-based prediction of the Saccharomyces cerevisiae SH3-ligand interactions.J Mol Biol. 2009 May 15;388(4):902-16. doi: 10.1016/j.jmb.2009.03.038. Epub 2009 Mar 24. J Mol Biol. 2009. PMID: 19324052
-
Binding constraints on the evolution of enzymes and signalling proteins: the important role of negative pleiotropy.Proc Biol Sci. 2011 Jul 7;278(1714):1930-5. doi: 10.1098/rspb.2010.2637. Epub 2011 Apr 13. Proc Biol Sci. 2011. PMID: 21490020 Free PMC article. Review.
-
Evolutionary predictions of binding surfaces and interactions.Curr Opin Struct Biol. 2002 Feb;12(1):21-7. doi: 10.1016/s0959-440x(02)00284-1. Curr Opin Struct Biol. 2002. PMID: 11839485 Review.
Cited by
-
Structural and biochemical analyses of selectivity determinants in chimeric Streptococcus Class A sortase enzymes.Protein Sci. 2022 Mar;31(3):701-715. doi: 10.1002/pro.4266. Epub 2022 Jan 3. Protein Sci. 2022. PMID: 34939250 Free PMC article.
-
Site-specific 2D IR spectroscopy: a general approach for the characterization of protein dynamics with high spatial and temporal resolution.Phys Chem Chem Phys. 2019 Jan 2;21(2):780-788. doi: 10.1039/c8cp06146g. Phys Chem Chem Phys. 2019. PMID: 30548035 Free PMC article.
-
A disordered encounter complex is central to the yeast Abp1p SH3 domain binding pathway.PLoS Comput Biol. 2020 Sep 14;16(9):e1007815. doi: 10.1371/journal.pcbi.1007815. eCollection 2020 Sep. PLoS Comput Biol. 2020. PMID: 32925900 Free PMC article.
-
Targeting Grb2 SH3 Domains with Affimer Proteins Provides Novel Insights into Ras Signalling Modulation.Biomolecules. 2024 Aug 22;14(8):1040. doi: 10.3390/biom14081040. Biomolecules. 2024. PMID: 39199427 Free PMC article.
-
Dissection of the role of a Src homology 3 domain in the evolution of binding preference of paralogous proteins.Genetics. 2024 Jan 3;226(1):iyad175. doi: 10.1093/genetics/iyad175. Genetics. 2024. PMID: 37793087 Free PMC article.
References
-
- Jen-Jacobson L. Protein-DNA recognition complexes: conservation of structure and binding energy in the transition state. Biopolymers. 1997;44(2):153–80. doi: 10.1002/(SICI)1097-0282(1997)44:2%3C153::AID-BIP4%3E3.0.CO;2-U - DOI - PubMed
-
- Takeda Y, Sarai A, Rivera VM. Analysis of the sequence-specific interactions between Cro repressor and operator DNA by systematic base substitution experiments. Proc Natl Acad Sci U S A. 1989;86(2):439–43. doi: 10.1073/pnas.86.2.439 - DOI - PMC - PubMed
-
- Greenspan NS. Cohen’s Conjecture, Howard’s Hypothesis, and Ptashne’s Ptruth: an exploration of the relationship between affinity and specificity. Trends in immunology. 2010;31(4):138–143. doi: 10.1016/j.it.2010.01.001 - DOI - PubMed
-
- Schreiber G, Keating AE. Protein binding specificity versus promiscuity. Current opinion in structural biology. 2011;21(1):50–61. doi: 10.1016/j.sbi.2010.10.002 - DOI - PMC - PubMed
-
- Szwajkajzer D, Carey J. Molecular and biological constraints on ligand-binding affinity and specificity. Biopolymers. 1997;44(2):181–98. doi: 10.1002/(SICI)1097-0282(1997)44:2%3C181::AID-BIP5%3E3.0.CO;2-R - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

