High-quality human and rat spermatozoal RNA isolation for functional genomic studies
- PMID: 29470852
- PMCID: PMC6309170
- DOI: 10.1111/andr.12471
High-quality human and rat spermatozoal RNA isolation for functional genomic studies
Abstract
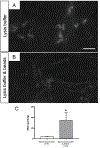
Sperm RNA is a sensitive monitoring endpoint for male reproductive toxicants, and a potential biomarker to assess male infertility and sperm quality. However, isolation of sperm RNA is a challenging procedure due to the heterogeneous population of cells present in the ejaculate, the low yield of RNA per spermatozoon, and the absence of 18S and 28S ribosomal RNA subunits. The unique biology of spermatozoa has created some uncertainty in the field about RNA isolation methods, indicating the need for rigorous quality control checks to ensure reproducibility of data generated from sperm RNA. Therefore, we developed a reliable and effective protocol for RNA isolation from rat and human spermatozoa that delivers highly purified and intact RNA, verified using RNA-specific electrophoretic chips and molecular biology approaches such as RT-PCR and Western blot analysis. The sperm RNA isolation technique was optimized using rat spermatozoa and then adapted to human spermatozoa. Three steps in the sperm isolation procedure, epididymal fluid collection, sperm purification, and spermatozoon RNA extraction, were evaluated and assessed. The sperm RNA extraction methodology consists of collection of rat epididymal fluid with repeated needle punctures of the epididymis, somatic cell elimination using detergent-based somatic cell lysis buffer (SCLB) and the use of RNA isolation Kit. Rat sperm heads are more resistant to disruption than human spermatozoa, necessitating the addition of mechanical lysis with microbeads and heat in the rat protocol, whereas the human sperm protocol only required lysis buffer. In conclusion, this methodology results in reliable and consistent isolation of high-quality sperm RNA. Using this technique will aid in translation of data collected from animal models, and reproducibility of clinical assessment of male factor fertility using RNA molecular biomarkers.
Keywords: RNA; high-quality methodology; human; integrity; rat; spermatozoa.
© 2018 American Society of Andrology and European Academy of Andrology.
Figures





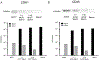
References
-
- Inhorn MC, Patrizio P. Infertility around the globe: new thinking on gender, reproductive technologies and global movements in the 21st century. Hum Reprod Update 2015; 21: 411–26. - PubMed
-
- Jarow JP, Sharlip ID, Belker AM, Lipshultz LI, Sigman M, Thomas AJ, et al. Best practice policies for male infertility. J Urol 2002; 167: 2138–44. - PubMed
-
- Gottardo F, Kliesch S, World Health O. [Semen analysis: spermiogram according to WHO 2010 criteria]. Urologe A 2011; 50: 101–8. - PubMed
-
- Kierszenbaum AL. Mammalian spermatogenesis in vivo and in vitro: a partnership of spermatogenic and somatic cell lineages. Endocr Rev 1994; 15: 116–34. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

