Integrator subunit 4 is a 'Symplekin-like' scaffold that associates with INTS9/11 to form the Integrator cleavage module
- PMID: 29471365
- PMCID: PMC5934644
- DOI: 10.1093/nar/gky100
Integrator subunit 4 is a 'Symplekin-like' scaffold that associates with INTS9/11 to form the Integrator cleavage module
Abstract
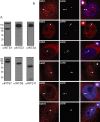
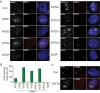
Integrator (INT) is a transcriptional regulatory complex associated with RNA polymerase II that is required for the 3'-end processing of both UsnRNAs and enhancer RNAs. Integrator subunits 9 (INTS9) and INTS11 constitute the catalytic core of INT and are paralogues of the cleavage and polyadenylation specificity factors CPSF100 and CPSF73. While CPSF73/100 are known to associate with a third protein called Symplekin, there is no paralog of Symplekin within INT raising the question of how INTS9/11 associate with the other INT subunits. Here, we have identified that INTS4 is a specific and conserved interaction partner of INTS9/11 that does not interact with either subunit individually. Although INTS4 has no significant homology with Symplekin, it possesses N-terminal HEAT repeats similar to Symplekin but also contains a β-sheet rich C-terminal region, both of which are important to bind INTS9/11. We assess three functions of INT including UsnRNA 3'-end processing, maintenance of Cajal body structural integrity, and formation of histone locus bodies to conclude that INTS4/9/11 are the most critical of the INT subunits for UsnRNA biogenesis. Altogether, these results indicate that INTS4/9/11 compose a heterotrimeric complex that likely represents the Integrator 'cleavage module' responsible for its endonucleolytic activity.
Figures






References
-
- Baillat D., Hakimi M.A., Naar A.M., Shilatifard A., Cooch N., Shiekhattar R.. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005; 123:265–276. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

