The H-subunit of the restriction endonuclease CglI contains a prototype DEAD-Z1 helicase-like motor
- PMID: 29471489
- PMCID: PMC5861437
- DOI: 10.1093/nar/gky107
The H-subunit of the restriction endonuclease CglI contains a prototype DEAD-Z1 helicase-like motor
Abstract
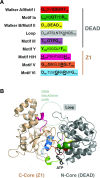
CglI is a restriction endonuclease from Corynebacterium glutamicum that forms a complex between: two R-subunits that have site specific-recognition and nuclease domains; and two H-subunits, with Superfamily 2 helicase-like DEAD domains, and uncharacterized Z1 and C-terminal domains. ATP hydrolysis by the H-subunits catalyses dsDNA translocation that is necessary for long-range movement along DNA that activates nuclease activity. Here, we provide biochemical and molecular modelling evidence that shows that Z1 has a fold distantly-related to RecA, and that the DEAD-Z1 domains together form an ATP binding interface and are the prototype of a previously undescribed monomeric helicase-like motor. The DEAD-Z1 motor has unusual Walker A and Motif VI sequences those nonetheless have their expected functions. Additionally, it contains DEAD-Z1-specific features: an H/H motif and a loop (aa 163-aa 172), that both play a role in the coupling of ATP hydrolysis to DNA cleavage. We also solved the crystal structure of the C-terminal domain which has a unique fold, and demonstrate that the Z1-C domains are the principal DNA binding interface of the H-subunit. Finally, we use small angle X-ray scattering to provide a model for how the H-subunit domains are arranged in a dimeric complex.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

