Single nucleotide polymorphisms in native South American Atlantic coast populations of smooth shelled mussels: hybridization with invasive European Mytilus galloprovincialis
- PMID: 29471805
- PMCID: PMC5824471
- DOI: 10.1186/s12711-018-0376-z
Single nucleotide polymorphisms in native South American Atlantic coast populations of smooth shelled mussels: hybridization with invasive European Mytilus galloprovincialis
Abstract
Background: Throughout the world, harvesting of mussels Mytilus spp. is based on the exploitation of natural populations and aquaculture. Aquaculture activities include transfers of spat and live adult mussels between various geographic locations, which may result in large-scale changes in the world distribution of Mytilus taxa. Mytilus taxa are morphologically similar and difficult to distinguish. In spite of much research on taxonomy, evolution and geographic distribution, the native Mytilus taxa of the Southern Hemisphere are poorly understood. Recently, single nucleotide polymorphisms (SNPs) have been used to clarify the taxonomic status of populations of smooth shelled mussels from the Pacific coast of South America. In this paper, we used a set of SNPs to characterize, for the first time, populations of smooth shelled mussels Mytilus from the Atlantic coast of South America.
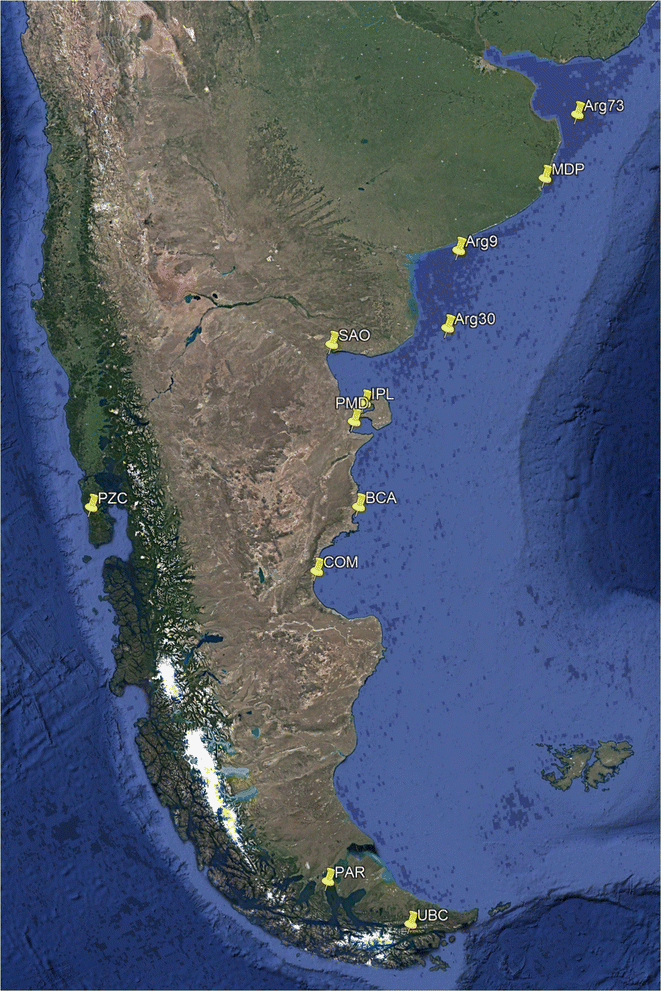
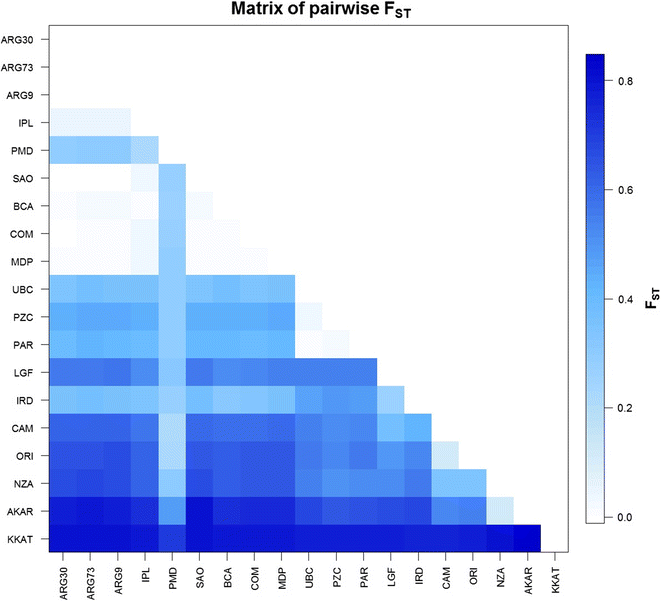
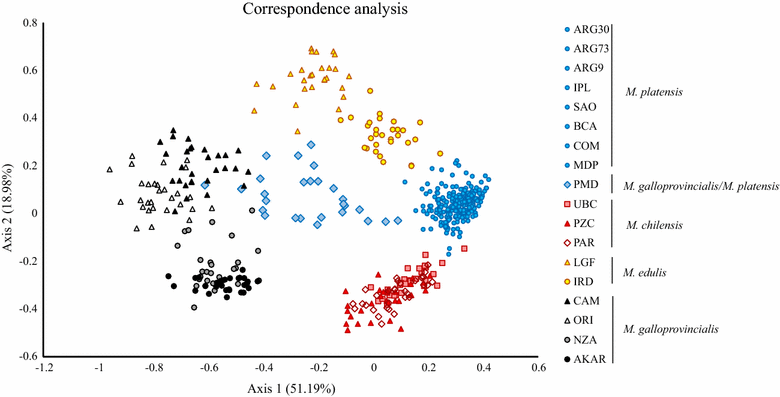
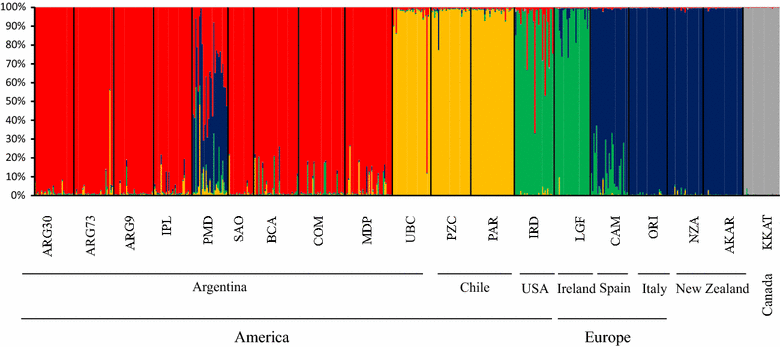
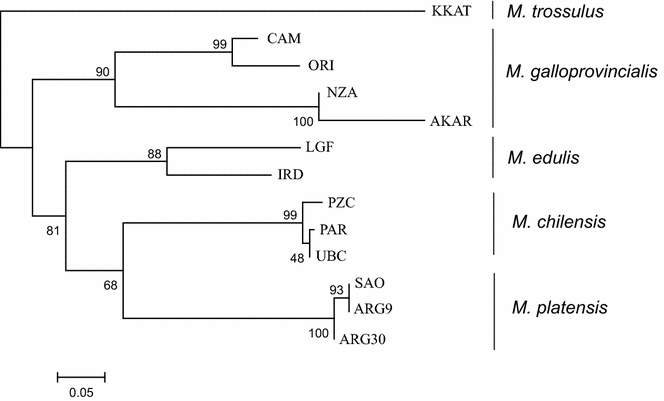
Results: Mytilus spp. samples were collected from eastern South America. Six reference samples from the Northern Hemisphere were used: Mytilus edulis from USA and Northern Ireland, Mytilus trossulus from Canada, and Mytilus galloprovincialis from Spain and Italy. Two other reference samples from the Southern Hemisphere were included: M. galloprovincialis from New Zealand and Mytilus chilensis from Chile. Fifty-five SNPs were successfully genotyped, of which 51 were polymorphic. Population genetic analyses using the STRUCTURE program revealed the clustering of eight populations from Argentina (Mytilus platensis) and the clustering of the sample from Ushuaia with M. chilensis from Chile. All individuals in the Puerto Madryn (Argentina) sample were identified as M. platensis × M. galloprovincialis F2 (88.89%) hybrids, except one that was classified as Mediterranean M. galloprovincialis. No F1 hybrids were observed.
Conclusions: We demonstrate that M. platensis (or Mytilus edulis platensis) and M. chilensis are distinct native taxa in South America, which indicates that the evolutionary histories of Mytilus taxa along the Atlantic and Pacific coasts differ. M. platensis is endangered by hybridization with M. galloprovincialis that was introduced from Europe into the Puerto Madryn area in Argentina, presumably by accidental introduction via ship traffic. We confirm the occurrence of a native M. chilensis population in southern Argentina on the coast of Patagonia.
Figures
Similar articles
-
Genetics and taxonomy of Chilean smooth-shelled mussels, Mytilus spp. (Bivalvia: Mytilidae).C R Biol. 2012 Jan;335(1):51-61. doi: 10.1016/j.crvi.2011.10.002. Epub 2011 Nov 4. C R Biol. 2012. PMID: 22226163
-
Pleistocene separation of mitochondrial lineages of Mytilus spp. mussels from Northern and Southern Hemispheres and strong genetic differentiation among southern populations.Mol Phylogenet Evol. 2008 Oct;49(1):84-91. doi: 10.1016/j.ympev.2008.07.006. Epub 2008 Jul 17. Mol Phylogenet Evol. 2008. PMID: 18678263
-
Blue mussels of the Mytilus edulis species complex from South America: The application of species delimitation models to DNA sequence variation.PLoS One. 2021 Sep 2;16(9):e0256961. doi: 10.1371/journal.pone.0256961. eCollection 2021. PLoS One. 2021. PMID: 34473778 Free PMC article.
-
Local adaptation and species segregation in two mussel (Mytilus edulis x Mytilus trossulus) hybrid zones.Mol Ecol. 2005 Feb;14(2):381-400. doi: 10.1111/j.1365-294X.2004.02379.x. Mol Ecol. 2005. PMID: 15660932 Review.
-
Mytilus hybridisation and impact on aquaculture: A minireview.Mar Genomics. 2016 Jun;27:3-7. doi: 10.1016/j.margen.2016.04.008. Epub 2016 May 2. Mar Genomics. 2016. PMID: 27157133 Review.
Cited by
-
The Mediterranean Mussel Mytilus galloprovincialis (Mollusca: Bivalvia) in Chile: Distribution and Genetic Structure of a Recently Introduced Invasive Marine Species.Animals (Basel). 2024 Mar 7;14(6):823. doi: 10.3390/ani14060823. Animals (Basel). 2024. PMID: 38539921 Free PMC article.
-
Unveiling the Impact of Gene Presence/Absence Variation in Driving Inter-Individual Sequence Diversity within the CRP-I Gene Family in Mytilus spp.Genes (Basel). 2023 Mar 24;14(4):787. doi: 10.3390/genes14040787. Genes (Basel). 2023. PMID: 37107545 Free PMC article.
-
Trans-Atlantic Distribution and Introgression as Inferred from Single Nucleotide Polymorphism: Mussels Mytilus and Environmental Factors.Genes (Basel). 2020 May 10;11(5):530. doi: 10.3390/genes11050530. Genes (Basel). 2020. PMID: 32397617 Free PMC article.
-
Structure, Evolution, and Mitochondrial Genome Analysis of Mussel Species (Bivalvia, Mytilidae).Int J Mol Sci. 2024 Jun 24;25(13):6902. doi: 10.3390/ijms25136902. Int J Mol Sci. 2024. PMID: 39000014 Free PMC article.
-
Some Examples of the Use of Molecular Markers for Needs of Basic Biology and Modern Society.Animals (Basel). 2021 May 20;11(5):1473. doi: 10.3390/ani11051473. Animals (Basel). 2021. PMID: 34065552 Free PMC article. Review.
References
-
- Kamermans P, Galley T, Boudry P, Fuentes J, McCombie H, Batista FM, et al. Blue mussel hatchery technology in Europe. In: Allan G, Burnell G, et al., editors. Advances in aquaculture hatchery technology. Oxford: Woodhead Publishing; 2013. pp. 339–373.
-
- FAO. Fisheries and Aquaculture Department. Cultured aquatic species information programme: Mytilus edulis (Linnaeus, 1758). http://www.fao.org/fishery/culturedspecies/Mytilus_edulis/en#tcNA00D6. Accessed 22 Jan 2018.
-
- Kijewski T, Wijsman JWM, Hummel H, Wenne R. Genetic composition of cultured and wild mussels Mytilus from The Netherlands and transfers from Ireland and Great Britain. Aquaculture. 2009;287:292–296. doi: 10.1016/j.aquaculture.2008.10.048. - DOI
-
- Smaal AC. European mussel cultivation along the Atlantic coast: production status, problems and perspectives. Hydrobiologia. 2002;484:89–98. doi: 10.1023/A:1021352904712. - DOI
-
- Muehlbauer F, Fraser D, Brenner M, Van Nieuwenhove K, Buck BH, Strand O, et al. Bivalve aquaculture transfers in Atlantic Europe. Part A: Transfer activities and legal framework. Ocean Coast Manag. 2014;89:127–138. doi: 10.1016/j.ocecoaman.2013.12.003. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

