Oncoprotein HBXIP enhances HOXB13 acetylation and co-activates HOXB13 to confer tamoxifen resistance in breast cancer
- PMID: 29471853
- PMCID: PMC5824486
- DOI: 10.1186/s13045-018-0577-5
Oncoprotein HBXIP enhances HOXB13 acetylation and co-activates HOXB13 to confer tamoxifen resistance in breast cancer
Abstract
Background: Resistance to tamoxifen (TAM) frequently occurs in the treatment of estrogen receptor positive (ER+) breast cancer. Accumulating evidences indicate that transcription factor HOXB13 is of great significance in TAM resistance. However, the regulation of HOXB13 in TAM-resistant breast cancer remains largely unexplored. Here, we were interested in the potential effect of HBXIP, an oncoprotein involved in the acceleration of cancer progression, on the modulation of HOXB13 in TAM resistance of breast cancer.
Methods: The Kaplan-Meier plotter cancer database and GEO dataset were used to analyze the association between HBXIP expression and relapse-free survival. The correlation of HBXIP and HOXB13 in ER+ breast cancer was assessed by human tissue microarray. Immunoblotting analysis, qRT-PCR assay, immunofluorescence staining, Co-IP assay, ChIP assay, luciferase reporter gene assay, cell viability assay, and colony formation assay were performed to explore the possible molecular mechanism by which HBXIP modulates HOXB13. Cell viability assay, xenograft assay, and immunohistochemistry staining analysis were utilized to evaluate the effect of the HBXIP/HOXB13 axis on the facilitation of TAM resistance in vitro and in vivo.
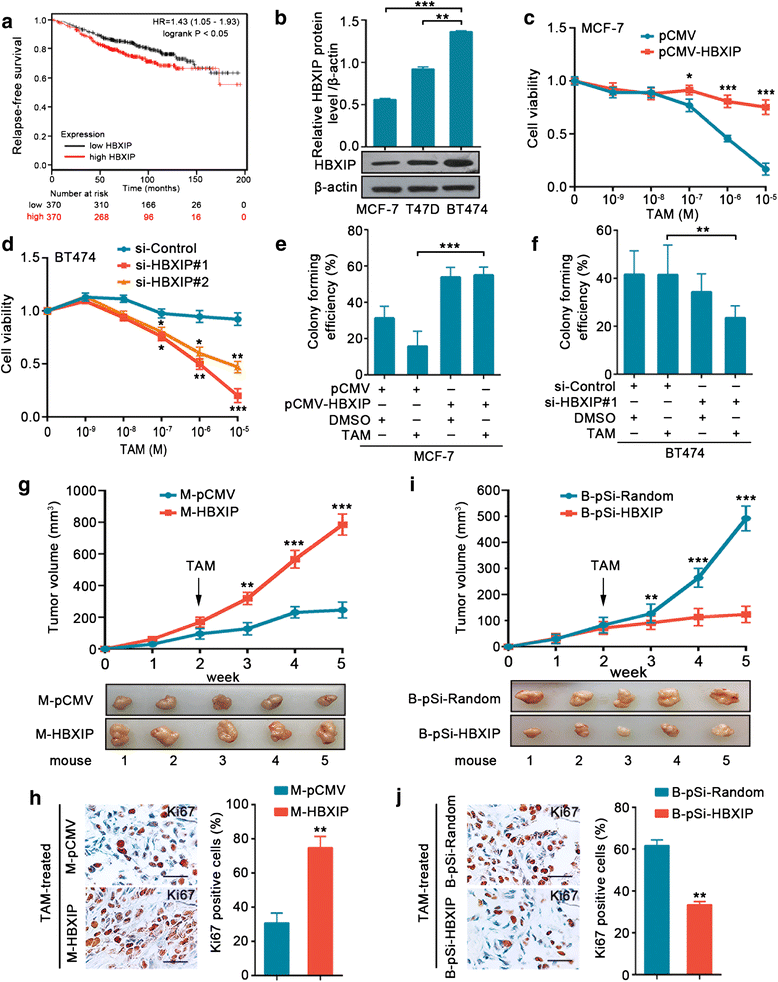
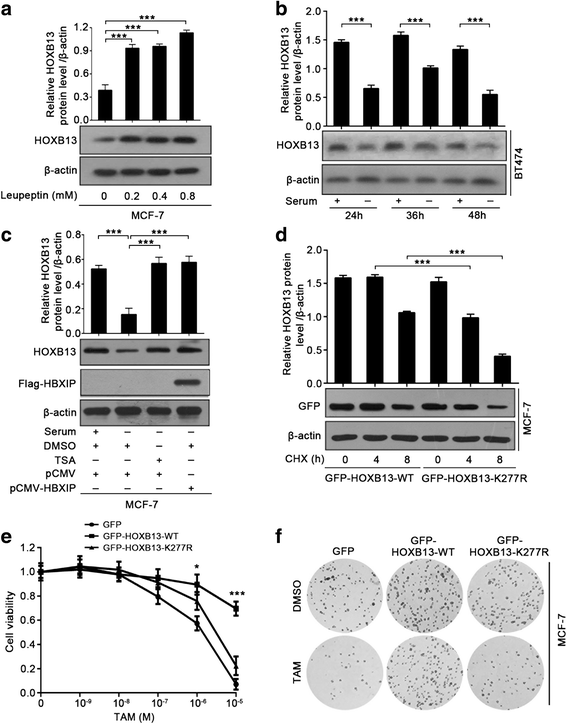
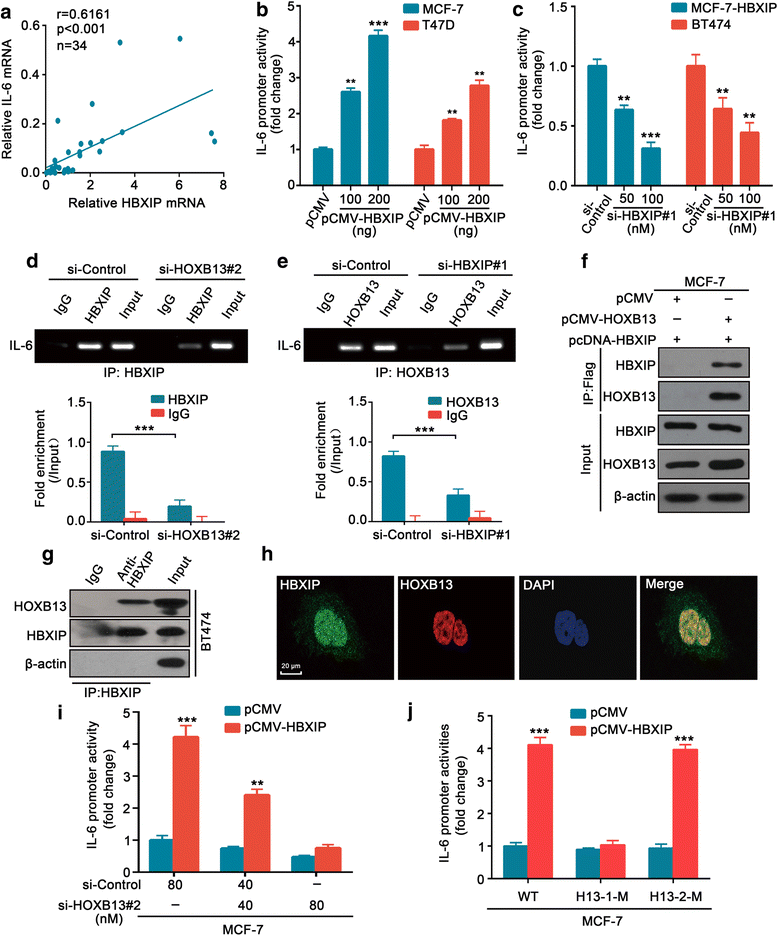
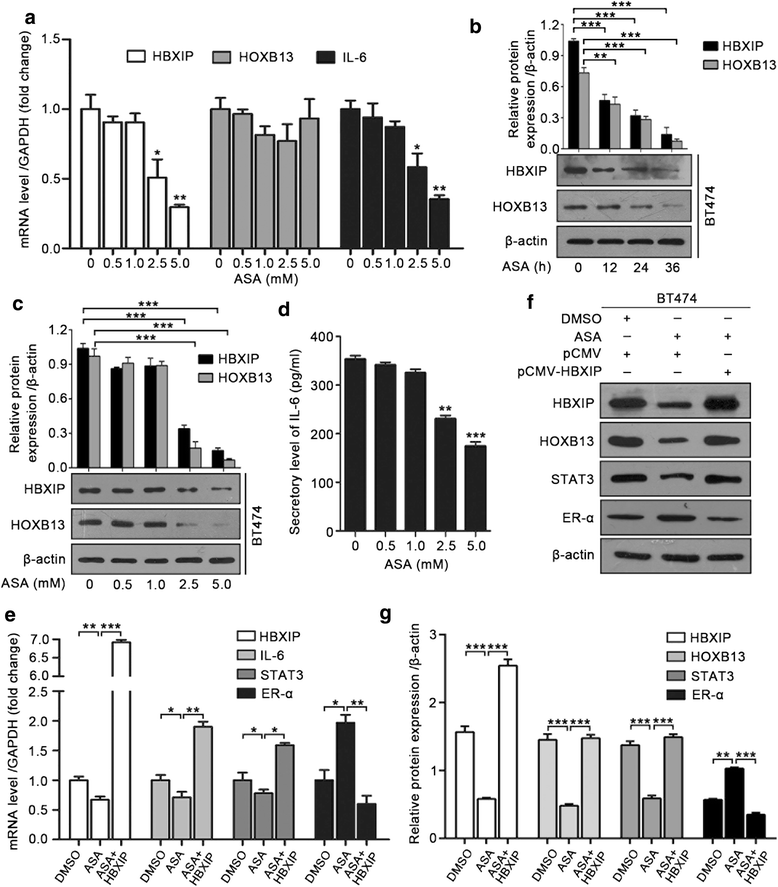
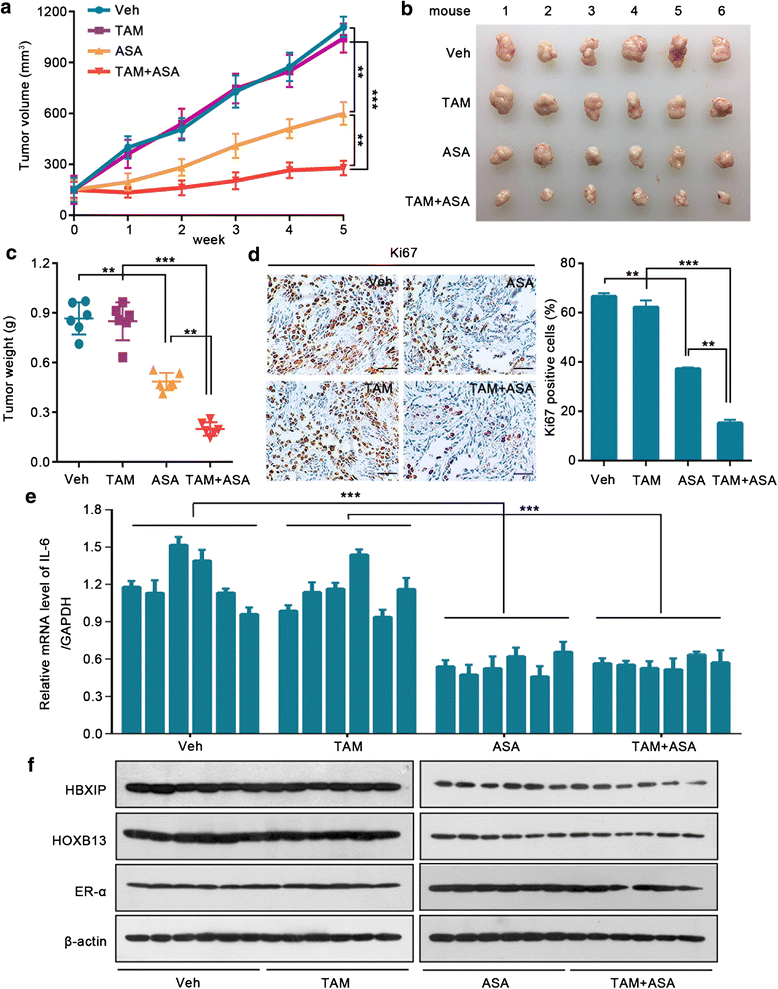
Results: The analysis of the Kaplan-Meier plotter and the GEO dataset showed that mono-TAM-treated breast cancer patients with higher HBXIP expression levels had shorter relapse-free survivals than patients with lower HBXIP expression levels. Overexpression of HBXIP induced TAM resistance in ER+ breast cancer cells. The tissue microarray analysis revealed a positive association between the expression levels of HBXIP and HOXB13 in ER+ breast cancer patients. HBXIP elevated HOXB13 protein level in breast cancer cells. Mechanistically, HBXIP prevented chaperone-mediated autophagy (CMA)-dependent degradation of HOXB13 via enhancement of HOXB13 acetylation at the lysine 277 residue, causing the accumulation of HOXB13. Moreover, HBXIP was able to act as a co-activator of HOXB13 to stimulate interleukin (IL)-6 transcription in the promotion of TAM resistance. Interestingly, aspirin (ASA) suppressed the HBXIP/HOXB13 axis by decreasing HBXIP expression, overcoming TAM resistance in vitro and in vivo.
Conclusions: Our study highlights that HBXIP enhances HOXB13 acetylation to prevent HOXB13 degradation and co-activates HOXB13 in the promotion of TAM resistance of breast cancer. Therapeutically, ASA can serve as a potential candidate for reversing TAM resistance by inhibiting HBXIP expression.
Keywords: Acetylation; Breast cancer; Chaperone-mediated autophagy; HBXIP; HOXB13; Tamoxifen resistance.
Conflict of interest statement
Ethics approval and consent to participate
This study was approved by the Research Ethics Committee of Nankai University, and written informed consent was obtained from all of the patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


Similar articles
-
HOXB13 mediates tamoxifen resistance and invasiveness in human breast cancer by suppressing ERα and inducing IL-6 expression.Cancer Res. 2013 Sep 1;73(17):5449-58. doi: 10.1158/0008-5472.CAN-13-1178. Epub 2013 Jul 5. Cancer Res. 2013. PMID: 23832664 Free PMC article.
-
HBXIP is a novel regulator of the unfolded protein response that sustains tamoxifen resistance in ER+ breast cancer.J Biol Chem. 2022 Mar;298(3):101644. doi: 10.1016/j.jbc.2022.101644. Epub 2022 Jan 28. J Biol Chem. 2022. PMID: 35093383 Free PMC article.
-
Oncoprotein HBXIP induces PKM2 via transcription factor E2F1 to promote cell proliferation in ER-positive breast cancer.Acta Pharmacol Sin. 2019 Apr;40(4):530-538. doi: 10.1038/s41401-018-0015-9. Epub 2018 Jun 20. Acta Pharmacol Sin. 2019. PMID: 29925919 Free PMC article.
-
[Cytotoxicity of tamoxifen and its principal metabolites in human breast cancer cell lines].Bull Cancer. 1996 Oct;83(10):808-15. Bull Cancer. 1996. PMID: 8952630 Review. French.
-
Targeting autophagy with tamoxifen in breast cancer: From molecular mechanisms to targeted therapy.Fundam Clin Pharmacol. 2023 Dec;37(6):1092-1108. doi: 10.1111/fcp.12936. Epub 2023 Jul 4. Fundam Clin Pharmacol. 2023. PMID: 37402635 Review.
Cited by
-
FTO demethylates m6A modifications in HOXB13 mRNA and promotes endometrial cancer metastasis by activating the WNT signalling pathway.RNA Biol. 2021 Sep;18(9):1265-1278. doi: 10.1080/15476286.2020.1841458. Epub 2020 Nov 5. RNA Biol. 2021. PMID: 33103587 Free PMC article.
-
Unconventional protein post-translational modifications: the helmsmen in breast cancer.Cell Biosci. 2022 Feb 25;12(1):22. doi: 10.1186/s13578-022-00756-z. Cell Biosci. 2022. PMID: 35216622 Free PMC article. Review.
-
Prognostic value of chronic hepatitis B virus infection in patients with breast cancer in a hepatitis B virus endemic area.Ann Transl Med. 2020 Mar;8(5):180. doi: 10.21037/atm.2020.01.97. Ann Transl Med. 2020. PMID: 32309327 Free PMC article.
-
Compared Inhibitory Activities of Tamoxifen and Avenanthramide B on Liver Esterase and Correlation Based on the Superimposed Structure Between Porcine and Human Liver Esterase.Int J Mol Sci. 2024 Dec 11;25(24):13291. doi: 10.3390/ijms252413291. Int J Mol Sci. 2024. PMID: 39769055 Free PMC article.
-
The modulation of PD-L1 induced by the oncogenic HBXIP for breast cancer growth.Acta Pharmacol Sin. 2022 Feb;43(2):429-445. doi: 10.1038/s41401-021-00631-6. Epub 2021 Apr 6. Acta Pharmacol Sin. 2022. PMID: 33824459 Free PMC article.
References
-
- Connor CE, Norris JD, Broadwater G, Willson TM, Gottardis MM, Dewhirst MW, McDonnell DP. Circumventing tamoxifen resistance in breast cancers using antiestrogens that induce unique conformational changes in the estrogen receptor. Cancer Res. 2001;61:2917–2922. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

