A reservoir of 'historical' antibiotic resistance genes in remote pristine Antarctic soils
- PMID: 29471872
- PMCID: PMC5824556
- DOI: 10.1186/s40168-018-0424-5
A reservoir of 'historical' antibiotic resistance genes in remote pristine Antarctic soils
Abstract
Background: Soil bacteria naturally produce antibiotics as a competitive mechanism, with a concomitant evolution, and exchange by horizontal gene transfer, of a range of antibiotic resistance mechanisms. Surveys of bacterial resistance elements in edaphic systems have originated primarily from human-impacted environments, with relatively little information from remote and pristine environments, where the resistome may comprise the ancestral gene diversity.
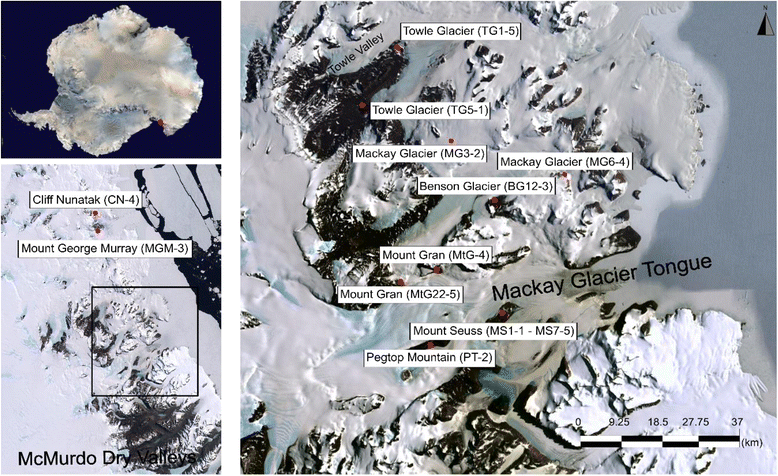
Methods: We used shotgun metagenomics to assess antibiotic resistance gene (ARG) distribution in 17 pristine and remote Antarctic surface soils within the undisturbed Mackay Glacier region. We also interrogated the phylogenetic placement of ARGs compared to environmental ARG sequences and tested for the presence of horizontal gene transfer elements flanking ARGs.
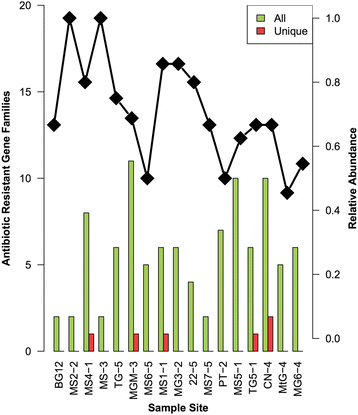
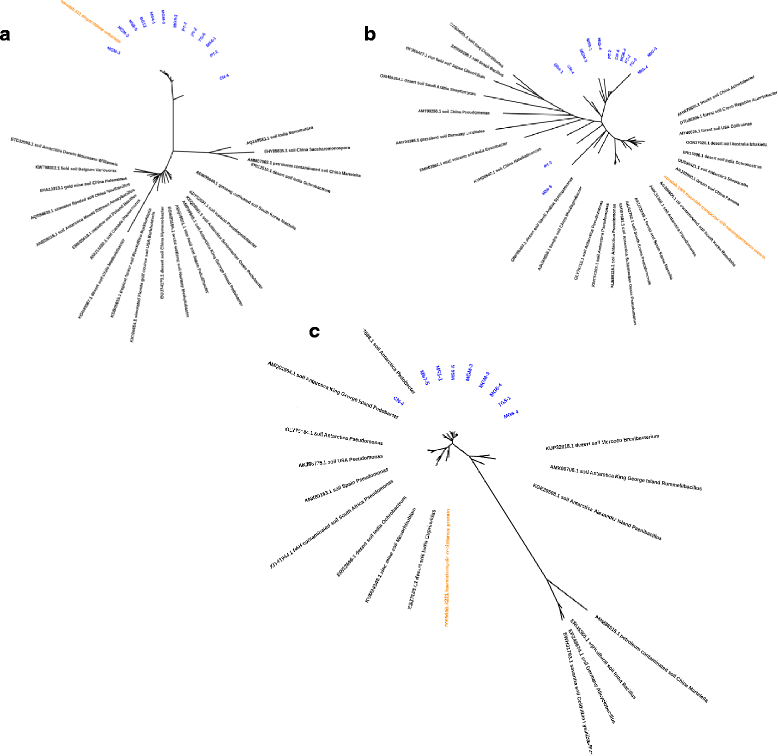
Results: In total, 177 naturally occurring ARGs were identified, most of which encoded single or multi-drug efflux pumps. Resistance mechanisms for the inactivation of aminoglycosides, chloramphenicol and β-lactam antibiotics were also common. Gram-negative bacteria harboured most ARGs (71%), with fewer genes from Gram-positive Actinobacteria and Bacilli (Firmicutes) (9%), reflecting the taxonomic composition of the soils. Strikingly, the abundance of ARGs per sample had a strong, negative correlation with species richness (r = - 0.49, P < 0.05). This result, coupled with a lack of mobile genetic elements flanking ARGs, suggests that these genes are ancient acquisitions of horizontal transfer events.
Conclusions: ARGs in these remote and uncontaminated soils most likely represent functional efficient historical genes that have since been vertically inherited over generations. The historical ARGs in these pristine environments carry a strong phylogenetic signal and form a monophyletic group relative to ARGs from other similar environments.
Keywords: Antarctica; Antibiotic resistance genes; Metagenomics; Soil resistome.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- WHO . Antimicrobial resistance: global report on surveillance 2014. Geneva: WHO; 2014.
-
- Cytryn E. The soil resistome: the anthropogenic, the native, and the unknown. Soil Biol Biochem. 2013;63:18–23. doi: 10.1016/j.soilbio.2013.03.017. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

