Modeling human pancreatic beta cell dedifferentiation
- PMID: 29472102
- PMCID: PMC5985229
- DOI: 10.1016/j.molmet.2018.02.002
Modeling human pancreatic beta cell dedifferentiation
Abstract
Objective: Dedifferentiation could explain reduced functional pancreatic β-cell mass in type 2 diabetes (T2D).
Methods: Here we model human β-cell dedifferentiation using growth factor stimulation in the human β-cell line, EndoC-βH1, and human pancreatic islets.
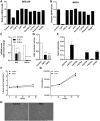
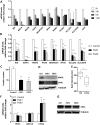
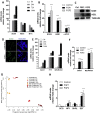
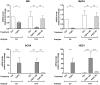
Results: Fibroblast growth factor 2 (FGF2) treatment reduced expression of β-cell markers, (INS, MAFB, SLC2A2, SLC30A8, and GCK) and activated ectopic expression of MYC, HES1, SOX9, and NEUROG3. FGF2-induced dedifferentiation was time- and dose-dependent and reversible upon wash-out. Furthermore, FGF2 treatment induced expression of TNFRSF11B, a decoy receptor for RANKL and protected β-cells against RANKL signaling. Finally, analyses of transcriptomic data revealed increased FGF2 expression in ductal, endothelial, and stellate cells in pancreas from T2D patients, whereas FGFR1, SOX,9 and HES1 expression increased in islets from T2D patients.
Conclusions: We thus developed an FGF2-induced model of human β-cell dedifferentiation, identified new markers of dedifferentiation, and found evidence for increased pancreatic FGF2, FGFR1, and β-cell dedifferentiation in T2D.
Keywords: Beta-cell; Dedifferentiation; Human; Type 2 diabetes.
Copyright © 2018 The Authors. Published by Elsevier GmbH.. All rights reserved.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

