Systematic Complex Haploinsufficiency-Based Genetic Analysis of Candida albicans Transcription Factors: Tools and Applications to Virulence-Associated Phenotypes
- PMID: 29472308
- PMCID: PMC5873919
- DOI: 10.1534/g3.117.300515
Systematic Complex Haploinsufficiency-Based Genetic Analysis of Candida albicans Transcription Factors: Tools and Applications to Virulence-Associated Phenotypes
Abstract
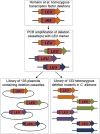
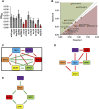
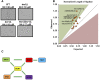
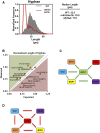
Genetic interaction analysis is a powerful approach to the study of complex biological processes that are dependent on multiple genes. Because of the largely diploid nature of the human fungal pathogen Candida albicans, genetic interaction analysis has been limited to a small number of large-scale screens and a handful for gene-by-gene studies. Complex haploinsufficiency, which occurs when a strain containing two heterozygous mutations at distinct loci shows a phenotype that is distinct from either of the corresponding single heterozygous mutants, is an expedient approach to genetic interactions analysis in diploid organisms. Here, we describe the construction of a barcoded-library of 133 heterozygous TF deletion mutants and deletion cassettes for designed to facilitate complex haploinsufficiency-based genetic interaction studies of the TF networks in C. albicans We have characterized the phenotypes of these heterozygous mutants under a broad range of in vitro conditions using both agar-plate and pooled signature tag-based assays. Consistent with previous studies, haploinsufficiency is relative uncommon. In contrast, a set of 12 TFs enriched in mutants with a role in adhesion were found to have altered competitive fitness at early time points in a murine model of disseminated candidiasis. Finally, we characterized the genetic interactions of a set of biofilm related TFs in the first two steps of biofilm formation, adherence and filamentation of adherent cells. The genetic interaction networks at each stage of biofilm formation are significantly different indicating that the network is not static but dynamic.
Keywords: Candida albicans; biofilm formation; complex haploinsufficiency; disseminated candidiasis; haploinsufficiency; hyphal morphogenesis.
Copyright © 2018 Glazier et al.
Figures





Similar articles
-
Construction of Double Heterozygous Deletion Strains for Complex Haploinsufficiency-Based Genetic Analysis in Candida albicans.Methods Mol Biol. 2022;2542:91-99. doi: 10.1007/978-1-0716-2549-1_6. Methods Mol Biol. 2022. PMID: 36008658
-
Genetic analysis of the Candida albicans biofilm transcription factor network using simple and complex haploinsufficiency.PLoS Genet. 2017 Aug 9;13(8):e1006948. doi: 10.1371/journal.pgen.1006948. eCollection 2017 Aug. PLoS Genet. 2017. PMID: 28793308 Free PMC article.
-
A large-scale complex haploinsufficiency-based genetic interaction screen in Candida albicans: analysis of the RAM network during morphogenesis.PLoS Genet. 2011 Apr;7(4):e1002058. doi: 10.1371/journal.pgen.1002058. PLoS Genet. 2011. PMID: 22103005 Free PMC article.
-
Genome-wide functional analysis in Candida albicans.Virulence. 2017 Nov 17;8(8):1563-1579. doi: 10.1080/21505594.2017.1292198. Epub 2017 Mar 13. Virulence. 2017. PMID: 28277904 Free PMC article. Review.
-
Candida albicans mutant construction and characterization of selected virulence determinants.J Microbiol Methods. 2015 Aug;115:153-65. doi: 10.1016/j.mimet.2015.06.004. Epub 2015 Jun 12. J Microbiol Methods. 2015. PMID: 26073905 Review.
Cited by
-
Construction of Double Heterozygous Deletion Strains for Complex Haploinsufficiency-Based Genetic Analysis in Candida albicans.Methods Mol Biol. 2022;2542:91-99. doi: 10.1007/978-1-0716-2549-1_6. Methods Mol Biol. 2022. PMID: 36008658
-
A novel genetic circuitry governing hypoxic metabolic flexibility, commensalism and virulence in the fungal pathogen Candida albicans.PLoS Pathog. 2019 Dec 6;15(12):e1007823. doi: 10.1371/journal.ppat.1007823. eCollection 2019 Dec. PLoS Pathog. 2019. PMID: 31809527 Free PMC article.
-
Genetic interaction analysis comes to the diploid human pathogen Candida albicans.PLoS Pathog. 2020 Apr 23;16(4):e1008399. doi: 10.1371/journal.ppat.1008399. eCollection 2020 Apr. PLoS Pathog. 2020. PMID: 32324799 Free PMC article. No abstract available.
-
Reinforcement amid genetic diversity in the Candida albicans biofilm regulatory network.PLoS Pathog. 2023 Jan 25;19(1):e1011109. doi: 10.1371/journal.ppat.1011109. eCollection 2023 Jan. PLoS Pathog. 2023. PMID: 36696432 Free PMC article.
-
The Candida albicans reference strain SC5314 contains a rare, dominant allele of the transcription factor Rob1 that modulates biofilm formation and oral commensalism.bioRxiv [Preprint]. 2023 Jun 17:2023.06.17.545405. doi: 10.1101/2023.06.17.545405. bioRxiv. 2023. Update in: mBio. 2023 Oct 31;14(5):e0152123. doi: 10.1128/mbio.01521-23. PMID: 37398495 Free PMC article. Updated. Preprint.
References
-
- Bharucha N., Chabrier-Rosello Y., Xu T., Johnson C., Sobczynski S., et al. , 2011. A large-scale complex haploinsufficiency-based genetic interaction screen in Candida albicans: analysis of the RAM network during morphogenesis. PLoS Genet. 7(4): e1002058 10.1371/journal.pgen.1002058 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
