Dynamic Control of Chromosome Topology and Gene Expression by a Chromatin Modification
- PMID: 29472317
- PMCID: PMC6041165
- DOI: 10.1101/sqb.2017.82.034439
Dynamic Control of Chromosome Topology and Gene Expression by a Chromatin Modification
Abstract
The function of chromatin modification in establishing higher-order chromosome structure during gene regulation has been elusive. We dissected the machinery and mechanism underlying the enrichment of histone modification H4K20me1 on hermaphrodite X chromosomes during Caenorhabditis elegans dosage compensation and discovered a key role for H4K20me1 in regulating X-chromosome topology and chromosome-wide gene expression. Structural and functional analysis of the dosage compensation complex (DCC) subunit DPY-21 revealed a novel Jumonji C demethylase subfamily that converts H4K20me2 to H4K20me1 in worms and mammals. Inactivation of demethylase activity in vivo by genome editing eliminated H4K20me1 enrichment on X chromosomes of somatic cells, increased X-linked gene expression, reduced X-chromosome compaction, and disrupted X-chromosome conformation by diminishing the formation of topologically associated domains. H4K20me1 is also enriched on the inactive X of female mice, making our studies directly relevant to mammalian development. Unexpectedly, DPY-21 also associates specifically with autosomes of nematode germ cells in a DCC-independent manner to enrich H4K20me1 and trigger chromosome compaction. Thus, DPY-21 is an adaptable chromatin regulator. Its H4K20me2 demethylase activity can be harnessed during development for distinct biological functions by targeting it to diverse genomic locations through different mechanisms. In both somatic cells and germ cells, H4K20me1 enrichment modulates three-dimensional chromosome architecture, demonstrating the direct link between chromatin modification and higher-order chromosome structure.
© 2017 Bian et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
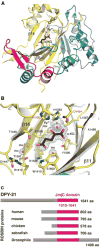
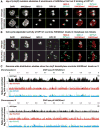



References
-
- Chen Z, Zang J, Whetstine J, Hong X, Davrazou F, Kutateladze TG, Simpson M, Mao Q, Pan CH, Dai S, et al. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125:691–702. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
