PIM1 mediates epithelial-mesenchymal transition by targeting Smads and c-Myc in the nucleus and potentiates clear-cell renal-cell carcinoma oncogenesis
- PMID: 29472550
- PMCID: PMC5833424
- DOI: 10.1038/s41419-018-0348-9
PIM1 mediates epithelial-mesenchymal transition by targeting Smads and c-Myc in the nucleus and potentiates clear-cell renal-cell carcinoma oncogenesis
Erratum in
-
Correction to: PIM1 mediates epithelial-mesenchymal transition by targeting Smads and c-Myc in the nucleus and potentiates clear-cell renal-cell carcinoma oncogenesis.Cell Death Dis. 2021 Mar 4;12(3):246. doi: 10.1038/s41419-021-03387-3. Cell Death Dis. 2021. PMID: 33664228 Free PMC article. No abstract available.
Abstract
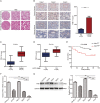
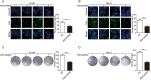
Emerging evidence has shown that the PIM serine/threonine kinase family, including PIM1, PIM2 and PIM3, is associated with tumour progression towards metastasis. PIM1, an attractive molecular target, has been identified as a potential prognostic biomarker for haematological and epithelial malignancies. However, to date, the potential regulatory roles and molecular mechanisms by which PIM1 affects the development and progression of cancers, including clear-cell renal-cell carcinoma (ccRCC), remain largely unknown. Herein, we present the first evidence that PIM1 is aberrantly overexpressed in human ccRCC tissues and cell lines and positively correlated with human ccRCC progression. In our study, depletion of PIM1 attenuated ccRCC cell proliferation, colony formation, migration, invasion and angiogenesis, suggesting that PIM1 expression may be a cancer-promoting event in ccRCC. Mechanistically, we observed that PIM1 could interact with Smad2 or Smad3 in the nucleus and subsequently phosphorylate Smad2 and Smad3 to induce the expression of transcription factors, including ZEB1, ZEB2, Snail1, Snail2 and Twist, to promote epithelial-mesenchymal transition (EMT). In addition, PIM1-mediated phosphorylation of c-Myc activates the expression of the above transcription factors to synergistically promote EMT but does not activate Smads. Collectively, our results demonstrate that aberrant expression of PIM1 contributes to ccRCC development and progression. Moreover, our data reveal a potential molecular mechanism in which PIM1 mediates crosstalk between signalling pathways, including different Smad proteins and c-Myc, which target downstream transcription factors (ZEB1, ZEB2, Snail1, Snail2 and Twist) to trigger EMT. Together, our data suggest that PIM1 may be a potential therapeutic target for ccRCC patients.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

