Heavy Chronic Intermittent Ethanol Exposure Alters Small Noncoding RNAs in Mouse Sperm and Epididymosomes
- PMID: 29472946
- PMCID: PMC5809758
- DOI: 10.3389/fgene.2018.00032
Heavy Chronic Intermittent Ethanol Exposure Alters Small Noncoding RNAs in Mouse Sperm and Epididymosomes
Abstract
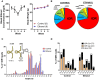
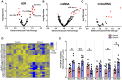
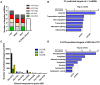
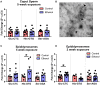
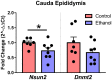
While the risks of maternal alcohol abuse during pregnancy are well-established, several preclinical studies suggest that chronic preconception alcohol consumption by either parent may also have significance consequences for offspring health and development. Notably, since isogenic male mice used in these studies are not involved in gestation or rearing of offspring, the cross-generational effects of paternal alcohol exposure suggest a germline-based epigenetic mechanism. Many recent studies have demonstrated that the effects of paternal environmental exposures such as stress or malnutrition can be transmitted to the next generation via alterations to small noncoding RNAs in sperm. Therefore, we used high throughput sequencing to examine the effect of preconception ethanol on small noncoding RNAs in sperm. We found that chronic intermittent ethanol exposure altered several small noncoding RNAs from three of the major small RNA classes in sperm, tRNA-derived small RNA (tDR), mitochondrial small RNA, and microRNA. Six of the ethanol-responsive small noncoding RNAs were evaluated with RT-qPCR on a separate cohort of mice and five of the six were confirmed to be altered by chronic ethanol exposure, supporting the validity of the sequencing results. In addition to altered sperm RNA abundance, chronic ethanol exposure affected post-transcriptional modifications to sperm small noncoding RNAs, increasing two nucleoside modifications previously identified in mitochondrial tRNA. Furthermore, we found that chronic ethanol reduced epididymal expression of a tRNA methyltransferase, Nsun2, known to directly regulate tDR biogenesis. Finally, ethanol-responsive sperm tDR are similarly altered in extracellular vesicles of the epididymis (i.e., epididymosomes), supporting the hypothesis that alterations to sperm tDR emerge in the epididymis and that epididymosomes are the primary source of small noncoding RNAs in sperm. These results add chronic ethanol to the growing list of paternal exposures that can affect small noncoding RNA abundance and nucleoside modifications in sperm. As small noncoding RNAs in sperm have been shown to causally induce heritable phenotypes in offspring, additional research is warranted to understand the potential effects of ethanol-responsive sperm small noncoding RNAs on offspring health and development.
Keywords: epididymosomes; epigenetics; ethanol; noncoding RNA; sperm.
Figures

References
-
- Benjamini Y., Kreiger A. M., Yekutieli D. (2006). Adaptive linear step-up procedures that control the false discovery rate. Biometrika 93, 491–507. 10.1093/biomet/93.3.491 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

