Codominant grasses differ in gene expression under experimental climate extremes in native tallgrass prairie
- PMID: 29473008
- PMCID: PMC5816582
- DOI: 10.7717/peerj.4394
Codominant grasses differ in gene expression under experimental climate extremes in native tallgrass prairie
Abstract
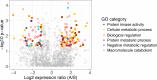
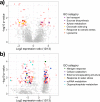
Extremes in climate, such as heat waves and drought, are expected to become more frequent and intense with forecasted climate change. Plant species will almost certainly differ in their responses to these stressors. We experimentally imposed a heat wave and drought in the tallgrass prairie ecosystem near Manhattan, Kansas, USA to assess transcriptional responses of two ecologically important C4 grass species, Andropogon gerardii and Sorghastrum nutans. Based on previous research, we expected that S. nutans would regulate more genes, particularly those related to stress response, under high heat and drought. Across all treatments, S. nutans showed greater expression of negative regulatory and catabolism genes while A. gerardii upregulated cellular and protein metabolism. As predicted, S. nutans showed greater sensitivity to water stress, particularly with downregulation of non-coding RNAs and upregulation of water stress and catabolism genes. A. gerardii was less sensitive to drought, although A. gerardii tended to respond with upregulation in response to drought versus S. nutans which downregulated more genes under drier conditions. Surprisingly, A. gerardii only showed minimal gene expression response to increased temperature, while S. nutans showed no response. Gene functional annotation suggested that these two species may respond to stress via different mechanisms. Specifically, A. gerardii tends to maintain molecular function while S. nutans prioritizes avoidance. Sorghastrum nutans may strategize abscisic acid response and catabolism to respond rapidly to stress. These results have important implications for success of these two important grass species under a more variable and extreme climate forecast for the future.
Keywords: Andropogon gerardii; C4 grass; Dominant species; Drought; Heat wave; Microarray; Sorghastrum nutans.
Conflict of interest statement
The authors declare there are no competing interests.
Figures

Similar articles
-
Gene expression differs in codominant prairie grasses under drought.Mol Ecol Resour. 2018 Mar;18(2):334-346. doi: 10.1111/1755-0998.12733. Epub 2017 Nov 20. Mol Ecol Resour. 2018. PMID: 29098789
-
Gene expression patterns of two dominant tallgrass prairie species differ in response to warming and altered precipitation.Sci Rep. 2016 May 13;6:25522. doi: 10.1038/srep25522. Sci Rep. 2016. PMID: 27174156 Free PMC article.
-
Barley Yellow Dwarf Disease in Natural Populations of Dominant Tallgrass Prairie Species in Kansas.Plant Dis. 2004 May;88(5):574. doi: 10.1094/PDIS.2004.88.5.574B. Plant Dis. 2004. PMID: 30812673
-
Photosynthetic responses of a dominant C4 grass to an experimental heat wave are mediated by soil moisture.Oecologia. 2017 Jan;183(1):303-313. doi: 10.1007/s00442-016-3755-6. Epub 2016 Oct 18. Oecologia. 2017. PMID: 27757543
-
Epigenomics in stress tolerance of plants under the climate change.Mol Biol Rep. 2023 Jul;50(7):6201-6216. doi: 10.1007/s11033-023-08539-6. Epub 2023 Jun 9. Mol Biol Rep. 2023. PMID: 37294468 Review.
Cited by
-
Climate variability supersedes grazing to determine the anatomy and physiology of a dominant grassland species.Oecologia. 2022 Feb;198(2):345-355. doi: 10.1007/s00442-022-05106-x. Epub 2022 Jan 12. Oecologia. 2022. PMID: 35018484 Free PMC article.
-
Flood-driven survival and growth of dominant C4 grasses helps set their distributions along tallgrass prairie moisture gradients.Am J Bot. 2025 Jan;112(1):e16457. doi: 10.1002/ajb2.16457. Epub 2025 Jan 8. Am J Bot. 2025. PMID: 39780373 Free PMC article.
References
-
- AbdElgawad H, Peshev D, Zinta G, Van Den Ende W, Janssens IA, Asard H. Climate extreme effects on the chemical composition of temperate grassland species under ambient and elevated CO2: a comparison of fructan and non-fructan accumulators. PLOS ONE. 2014;9(3):e92044. doi: 10.1371/journal.pone.0092044. - DOI - PMC - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

