Molecular analysis of integrons and antimicrobial resistance profile in Shigella spp. isolated from acute pediatric diarrhea patients
- PMID: 29473015
- PMCID: PMC5804674
- DOI: 10.3205/dgkh000308
Molecular analysis of integrons and antimicrobial resistance profile in Shigella spp. isolated from acute pediatric diarrhea patients
Abstract
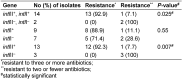
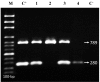
Introduction:Shigella spp. is a growing global health concern due to increasing multiple drug resistance, commonly resulting in therapeutic failure. Integrons are gene expression systems run by integrase genes. The aims of this study were detection of class I, II and III integrons and assessment of antimicrobial resistance in Shigella spp. isolated from acute pediatric diarrhea patients. Materials and methods: From January to December 2015, 16 Shigella spp. were isolated from 310 non-duplicative diarrheal stool samples in Children's Medical Center, Tehran, Iran. The isolates were analyzed for their antibiotic susceptibility using CLSI guidelines M100-S14. Multiplex PCR was used for amplification of I, II and III integron-associated integrase (intl) genes. Results: Of 310 stool samples, 16 (5.2%) were positive for Shigella spp., in 7 of them S. sonnei and in 9 of them S. flexneri were identified. Results of the antimicrobial susceptibility test showed that 6.2%, 50%, 31.2%, 6.2%, 81.2%, 56.2% and 31.2% of the isolates were resistant to gentamicin, chloramphenicol, nalidixic acid, ciprofloxacin, tetracycline, ampicillin and trimethoprim-sulfamethoxazole, respectively. Multiplex PCR results revealed that 6.2% (1/16), 31.2% (5/16), 50% (8/16) of Shigella isolates carried intlI, intlII and both intlI/intllI genes. No class 3 integrons were detected. Discussion: In this study, multidrug resistance was seen in Shigella isolates similar to that in isolates from other geographical areas. This is possible due to inappropriate use of antimicrobials. Furthermore, prevalence of multidrug resistance was significantly linked to the presence of integrin genes. Conclusion: A class 2 integron plays a role in presence of multidrug resistance in Shigella spp. It is vital to prevent the spread of antibiotic resistance through continuous monitoring.
Hintergrund: Die Zunahme multiresistenter Shigella spp. ist ein globales Gesundheitsproblem wachsender Bedeutung. Integrons sind Genexpressionssysteme, die von Integrase-Genen gesteuert werden. Zielsetzung der Studie war die Detektion von Klasse 1, 2 und 3 Integrons und die Bestimmung der antimikrobiellen Resistenz von Shigella spp., die von pädiatrischen Patienten mit Diarrhoe isoliert wurden. Material und Methoden: Von Januar bis Dezember 2015 wurden 16 Shigella spp. aus 310 nicht-duplikativen Durchfall-Stuhlproben im Children’s Medical Center, Tehran, gemäß Guideline des Clinical and Laboratory Standards Institute isoliert. Zur Amplifikation der I, II und III Integron-assoziierten Integrase(intl)-Gene wurde die Multiplex PCR eingesetzt.Ergebnisse: In 16 (5,2%) der 310 Stuhlproben wurden 7-mal S. sonnei und 9-mal S. flexneri isoliert. 6,2%, 50%, 31,2%, 6,2%, 81,2%, 56,2% bzw. 31,2% der Isolate waren resistent gegen Gentamicin, Chloramphenicol, Nalidixinsäure, Ciprofloxacin, Tetracycline, Ampicillin und Trimethoprim-Sulfamethoxazol. Mittels Multiplex PCR wurde nachgewiesen, dass 6,2% (1/16), 31,2% (5/16), 50% (8/16) der Shigella Isolate intlI, intlII bzw. beide Gene trugen. Klasse III Integrons wurden nicht detektiert.Diskussion: Bei Shigella-Isolaten wurde ähnlich zu anderen geographischen Regionen Multiresistenz nachgewiesen. Das wird begünstigt durch nicht Leitlinien gerechten Einsatz von Antibiotika. Die Prävalenz der Multiresistenz war signifikant mit dem Vorhandensein von Integrin-Genen assoziiert.Schlussfolgerung: Das Klasse 2 Integron ist von Bedeutung für die Multiresistenz von Shigella spp. Es ist wichtig, die Ausbreitung von Antibiotikaresistenzen durch kontinuierliche Überwachung zu verhindern.
Keywords: Shigella spp.; acute pediatric diarrhea; integrons; multiplex PCR.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Antimicrobial resistance patterns and prevalence of class 1 and 2 integrons in Shigella flexneri and Shigella sonnei isolated in Uzbekistan.Gut Pathog. 2010 Dec 9;2(1):18. doi: 10.1186/1757-4749-2-18. Gut Pathog. 2010. PMID: 21143880 Free PMC article.
-
Molecular epidemiology and genetic characterization of Shigella in pediatric patients in Iran.Infez Med. 2018 Dec 1;26(4):321-328. Infez Med. 2018. PMID: 30555135
-
Molecular characterization of multidrug-resistant Shigella spp. of food origin.Int J Food Microbiol. 2015 Feb 2;194:78-82. doi: 10.1016/j.ijfoodmicro.2014.11.013. Epub 2014 Nov 20. Int J Food Microbiol. 2015. PMID: 25485847
-
Prevalence of integron classes in Gram-negative clinical isolated bacteria in Iran: a systematic review and meta-analysis.Iran J Basic Med Sci. 2019 Feb;22(2):118-127. doi: 10.22038/ijbms.2018.32052.7697. Iran J Basic Med Sci. 2019. PMID: 30834075 Free PMC article. Review.
-
Prevalence of Antibiotic-Resistant Shigella spp. in Bangladesh: A Systematic Review and Meta-Analysis of 44,519 Samples.Antibiotics (Basel). 2023 Apr 26;12(5):817. doi: 10.3390/antibiotics12050817. Antibiotics (Basel). 2023. PMID: 37237720 Free PMC article. Review.
Cited by
-
Occurrence of shigellosis in pediatric diarrheal patients in Chattogram, Bangladesh: A molecular based approach.PLoS One. 2023 Jun 15;18(6):e0275353. doi: 10.1371/journal.pone.0275353. eCollection 2023. PLoS One. 2023. PMID: 37319254 Free PMC article.
-
Plasmids of Shigella flexneri serotype 1c strain Y394 provide advantages to bacteria in the host.BMC Microbiol. 2019 Apr 29;19(1):86. doi: 10.1186/s12866-019-1455-1. BMC Microbiol. 2019. PMID: 31035948 Free PMC article.
-
Fluoroquinolones-resistant Shigella species in Iranian children: a meta-analysis.World J Pediatr. 2019 Oct;15(5):441-453. doi: 10.1007/s12519-019-00263-1. Epub 2019 Jun 1. World J Pediatr. 2019. PMID: 31154582
-
Pharmacokinetics of tebipenem pivoxil used in children suffering from shigellosis: a pilot study in Bangladesh.Sci Rep. 2024 Dec 30;14(1):31965. doi: 10.1038/s41598-024-83549-3. Sci Rep. 2024. PMID: 39738375 Free PMC article. Clinical Trial.
-
The increasing antimicrobial resistance of Shigella species among Iranian pediatrics: a systematic review and meta-analysis.Pathog Glob Health. 2023 Oct;117(7):611-622. doi: 10.1080/20477724.2023.2179451. Epub 2023 Feb 16. Pathog Glob Health. 2023. PMID: 36794800 Free PMC article.
References
-
- Okeke IN, Laxminarayan R, Bhutta ZA, Duse AG, Jenkins P, O'Brien TF, Pablos-Mendez A, Klugman KP. Antimicrobial resistance in developing countries. Part I: recent trends and current status. Lancet Infect Dis. 2005 Aug;5(8):481–493. doi: 10.1016/S1473-3099(05)70189-4. doi: 10.1016/S1473-3099(05)70189-4. Available from: - DOI - DOI - PubMed
-
- Ye C, Lan R, Xia S, Zhang J, Sun Q, Zhang S, Jing H, Wang L, Li Z, Zhou Z, Zhao A, Cui Z, Cao J, Jin D, Huang L, Wang Y, Luo X, Bai X, Wang Y, Wang P, Xu Q, Xu J. Emergence of a new multidrug-resistant serotype X variant in an epidemic clone of Shigella flexneri. J Clin Microbiol. 2010 Feb;48(2):419–426. doi: 10.1128/JCM.00614-09. doi: 10.1128/JCM.00614-09. Available from: - DOI - DOI - PMC - PubMed
-
- Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012 Mar;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. doi: 10.1111/j.1469-0691.2011.03570.x. Available from: - DOI - DOI - PubMed
-
- Pan JC, Ye R, Meng DM, Zhang W, Wang HQ, Liu KZ. Molecular characteristics of class 1 and class 2 integrons and their relationships to antibiotic resistance in clinical isolates of Shigella sonnei and Shigella flexneri. J Antimicrob Chemother. 2006 Aug;58(2):288–296. doi: 10.1093/jac/dkl228. doi: 10.1093/jac/dkl228. Available from: - DOI - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

