Construction and characterization of bacterial artificial chromosomes harboring the full-length genome of a highly attenuated vaccinia virus LC16m8
- PMID: 29474493
- PMCID: PMC5825015
- DOI: 10.1371/journal.pone.0192725
Construction and characterization of bacterial artificial chromosomes harboring the full-length genome of a highly attenuated vaccinia virus LC16m8
Abstract
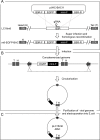
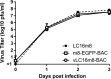
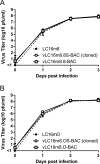
LC16m8 (m8), a highly attenuated vaccinia virus (VAC) strain, was developed as a smallpox vaccine, and its safety and immunogenicity have been confirmed. Here, we aimed to develop a system that recovers infectious m8 from a bacterial artificial chromosome (BAC) that retains the full-length viral genomic DNA (m8-BAC system). The infectious virus was successfully recovered from a VAC-BAC plasmid, named pLC16m8-BAC. Furthermore, the bacterial replicon-free virus was generated by intramolecular homologous recombination and was successfully recovered from a modified VAC-BAC plasmid, named pLC16m8.8S-BAC. Also, the growth of the recovered virus was indistinguishable from that of authentic m8. The full genome sequence of the plasmid, which harbors identical inverted terminal repeats (ITR) to that of authentic m8, was determined by long-read next-generation sequencing (NGS). The ITR contains x 18 to 32 of the 70 and x 30 to 45 of 54 base pair tandem repeats, and the number of tandem repeats was different between the ITR left and right. Since the virus recovered from pLC16m8.8S-BAC was expected to retain the identical viral genome to that of m8, including the ITR, a reference-based alignment following a short-read NGS was performed to validate the sequence of the recovered virus. Based on the pattern of coverage depth in the ITR, no remarkable differences were observed between the virus and m8, and the other region was confirmed to be identical as well. In summary, this new system can recover the virus, which is geno- and phenotypically indistinguishable from authentic m8.
Conflict of interest statement
Figures




Similar articles
-
An attenuated LC16m8 smallpox vaccine: analysis of full-genome sequence and induction of immune protection.J Virol. 2005 Sep;79(18):11873-91. doi: 10.1128/JVI.79.18.11873-11891.2005. J Virol. 2005. PMID: 16140764 Free PMC article.
-
Cloning the vaccinia virus genome as a bacterial artificial chromosome in Escherichia coli and recovery of infectious virus in mammalian cells.Proc Natl Acad Sci U S A. 2002 Sep 17;99(19):12415-20. doi: 10.1073/pnas.192420599. Epub 2002 Aug 26. Proc Natl Acad Sci U S A. 2002. PMID: 12196634 Free PMC article.
-
Cloning of the genomes of equine herpesvirus type 1 (EHV-1) strains KyA and racL11 as bacterial artificial chromosomes (BAC).J Vet Med B Infect Dis Vet Public Health. 2002 Feb;49(1):31-6. doi: 10.1046/j.1439-0450.2002.00534.x. J Vet Med B Infect Dis Vet Public Health. 2002. PMID: 11911590
-
The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses.Virology. 1998 May 10;244(2):365-96. doi: 10.1006/viro.1998.9123. Virology. 1998. PMID: 9601507 Review.
-
Development of viral infectious clones and their applications based on yeast and bacterial artificial chromosome platforms.Mol Biomed. 2025 Apr 29;6(1):26. doi: 10.1186/s43556-025-00266-7. Mol Biomed. 2025. PMID: 40295404 Free PMC article. Review.
Cited by
-
Markerless bacterial artificial chromosome manipulation method by red proteins of phage λ mediated homologous recombination utilizing fluorescent proteins for both positive and counter selection.Heliyon. 2023 Aug 7;9(8):e18983. doi: 10.1016/j.heliyon.2023.e18983. eCollection 2023 Aug. Heliyon. 2023. PMID: 37600421 Free PMC article.
-
Differences in the Likelihood of Acyclovir Resistance-Associated Mutations in the Thymidine Kinase Genes of Herpes Simplex Virus 1 and Varicella-Zoster Virus.Antimicrob Agents Chemother. 2019 Apr 25;63(5):e00017-19. doi: 10.1128/AAC.00017-19. Print 2019 May. Antimicrob Agents Chemother. 2019. PMID: 30858222 Free PMC article.
-
A Mutation in the Herpes Simplex Virus Type 1 (HSV-1) UL29 Gene is Associated with Anti-Herpesvirus Drugs' Susceptibility.Viruses. 2024 Nov 21;16(12):1813. doi: 10.3390/v16121813. Viruses. 2024. PMID: 39772124 Free PMC article.
-
Monkeypox: epidemiology, pathogenesis, treatment and prevention.Signal Transduct Target Ther. 2022 Nov 2;7(1):373. doi: 10.1038/s41392-022-01215-4. Signal Transduct Target Ther. 2022. PMID: 36319633 Free PMC article. Review.
References
-
- Aragon TJ, Ulrich S, Fernyak S, Rutherford GW. Risks of serious complications and death from smallpox vaccination: a systematic review of the United States experience, 1963–1968. BMC Public Health. 2003;3:26 doi: 10.1186/1471-2458-3-26 ; PubMed Central PMCID: PMCPMC194634. - DOI - PMC - PubMed
-
- Yamaguchi M, Kimura M, Hirayama M. Report of the National Smallpox Vaccination Research Committee: study of side effects, complications and their treatment. Clinical Virology. 1975;3:269–78.
-
- Morita M, Aoyama Y, Arita M, Amona H, Yoshizawa H, Hashizume S, et al. Comparative studies of several vaccinia virus strains by intrathalamic inoculation into cynomolgus monkeys. Arch Virol. 1977;53(3):197–208. . - PubMed
-
- Kenner J, Cameron F, Empig C, Jobes DV, Gurwith M. LC16m8: an attenuated smallpox vaccine. Vaccine. 2006;24(47–48):7009–22. doi: 10.1016/j.vaccine.2006.03.087 . - DOI - PMC - PubMed
-
- Morikawa S, Sakiyama T, Hasegawa H, Saijo M, Maeda A, Kurane I, et al. An attenuated LC16m8 smallpox vaccine: analysis of full-genome sequence and induction of immune protection. J Virol. 2005;79(18):11873–91. doi: 10.1128/JVI.79.18.11873-11891.2005 ; PubMed Central PMCID: PMCPMC1212643. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

