Time-Course Transcriptomics Analysis Reveals Key Responses of Submerged Deepwater Rice to Flooding
- PMID: 29475897
- PMCID: PMC5884608
- DOI: 10.1104/pp.17.00858
Time-Course Transcriptomics Analysis Reveals Key Responses of Submerged Deepwater Rice to Flooding
Abstract
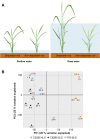
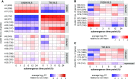
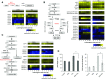
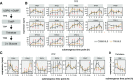
Water submergence is an environmental factor that limits plant growth and survival. Deepwater rice (Oryza sativa) adapts to submergence by rapidly elongating its internodes and thereby maintaining its leaves above the water surface. We performed a comparative RNA sequencing transcriptome analysis of the shoot base region, including basal nodes, internodes, and shoot apices of seedlings at two developmental stages from two varieties with contrasting deepwater growth responses. A transcriptomic comparison between deepwater rice cv C9285 and nondeepwater rice cv Taichung 65 revealed both similar and differential expression patterns between the two genotypes during submergence. The expression of genes related to gibberellin biosynthesis, trehalose biosynthesis, anaerobic fermentation, cell wall modification, and transcription factors that include ethylene-responsive factors was significantly different between the varieties. Interestingly, in both varieties, the jasmonic acid content at the shoot base decreased during submergence, while exogenous jasmonic acid inhibited submergence-induced internode elongation in cv C9285, suggesting that jasmonic acid plays a role in the submergence response of rice. Furthermore, a targeted de novo transcript assembly revealed transcripts that were specific to cv C9285, including submergence-induced biotic stress-related genes. Our multifaceted transcriptome approach using the rice shoot base region illustrates a differential response to submergence between deepwater and nondeepwater rice. Jasmonic acid metabolism appears to participate in the submergence-mediated internode elongation response of deepwater rice.
© 2018 American Society of Plant Biologists. All Rights Reserved.
Figures

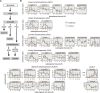
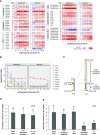
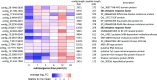
References
-
- Azuma T, Hirano T, Deki Y, Uchida N, Yasua T, Yamaguchi T (1995) Involvement of the decrease in levels of abscisic acid in the internodal elongation of submerged floating rice. J Plant Physiol 146: 323–328
-
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 - PubMed
-
- Bailey-Serres J, Voesenek LA (2008) Flooding stress: acclimations and genetic diversity. Annu Rev Plant Biol 59: 313–339 - PubMed
-
- Bailey-Serres J, Voesenek LA (2010) Life in the balance: a signaling network controlling survival of flooding. Curr Opin Plant Biol 13: 489–494 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

