Solution structure of the cytochrome P450 reductase-cytochrome c complex determined by neutron scattering
- PMID: 29475945
- PMCID: PMC5892573
- DOI: 10.1074/jbc.RA118.001941
Solution structure of the cytochrome P450 reductase-cytochrome c complex determined by neutron scattering
Abstract
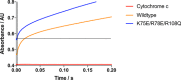
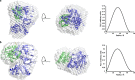
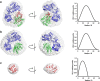
Electron transfer in all living organisms critically relies on formation of complexes between the proteins involved. The function of these complexes requires specificity of the interaction to allow for selective electron transfer but also a fast turnover of the complex, and they are therefore often transient in nature, making them challenging to study. Here, using small-angle neutron scattering with contrast matching with deuterated protein, we report the solution structure of the electron transfer complex between cytochrome P450 reductase (CPR) and its electron transfer partner cytochrome c This is the first reported solution structure of a complex between CPR and an electron transfer partner. The structure shows that the interprotein interface includes residues from both the FMN- and FAD-binding domains of CPR. In addition, the FMN is close to the heme of cytochrome c but distant from the FAD, indicating that domain movement is required between the electron transfer steps in the catalytic cycle of CPR. In summary, our results reveal key details of the CPR catalytic mechanism, including interactions of two domains of the reductase with cytochrome c and motions of these domains relative to one another. These findings shed light on interprotein electron transfer in this system and illustrate a powerful approach for studying solution structures of protein-protein complexes.
Keywords: SANS; cytochrome P450; cytochrome P450 reductase; cytochrome c; electron transfer; neutron scattering; protein structural dynamics; protein structure; protein-protein interaction; solution structure; transient complex.
© 2018 Freeman et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Ahuja S., Jahr N., Im S. C., Vivekanandan S., Popovych N., Le Clair S. V., Huang R., Soong R., Xu J., Yamamoto K., Nanga R. P., Bridges A., Waskell L., and Ramamoorthy A. (2013) A model of the membrane-bound cytochrome b5-cytochrome P450 complex from NMR and mutagenesis data. J. Biol. Chem. 288, 22080–22095 10.1074/jbc.M112.448225 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

