A chemical genetic approach to engineer phototropin kinases for substrate labeling
- PMID: 29475950
- PMCID: PMC5900772
- DOI: 10.1074/jbc.RA118.001834
A chemical genetic approach to engineer phototropin kinases for substrate labeling
Abstract
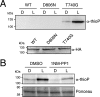
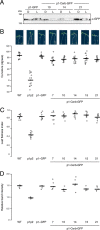
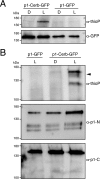
Protein kinases (PKs) control many aspects of plant physiology by regulating signaling networks through protein phosphorylation. Phototropins (phots) are plasma membrane-associated serine/threonine PKs that control a range of physiological processes that collectively serve to optimize photosynthetic efficiency in plants. These include phototropism, leaf positioning and flattening, chloroplast movement, and stomatal opening. Despite their identification over two decades ago, only a handful of substrates have been identified for these PKs. Progress in this area has been hampered by the lack of a convenient means to confirm the identity of potential substrate candidates. Here we demonstrate that the kinase domain of Arabidopsis phot1 and phot2 can be successfully engineered to accommodate non-natural ATP analogues by substituting the bulky gatekeeper residue threonine for glycine. This approach circumvents the need for radioactivity to track phot kinase activity and follow light-induced receptor autophosphorylation in vitro by incorporating thiophosphate from N6-benzyl-ATPγS. Consequently, thiophosphorylation of phot substrate candidates can be readily monitored when added or co-expressed with phots in vitro Furthermore, gatekeeper-modified phot1 retained its functionality and its ability to accommodate N6-benzyl-ATPγS as a phosphodonor when expressed in Arabidopsis We therefore anticipate that this chemical genetic approach will provide new opportunities for labeling and identifying substrates for phots and other related AGC kinases under in vitro and near-native in vivo conditions.
Keywords: ATP; Arabidopsis; chemical genetics; gatekeeper; photobiology; photoreceptor; protein engineering; protein kinase.
© 2018 Schnabel et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

