Highly multiplexed and quantitative cell-surface protein profiling using genetically barcoded antibodies
- PMID: 29476010
- PMCID: PMC5856557
- DOI: 10.1073/pnas.1721899115
Highly multiplexed and quantitative cell-surface protein profiling using genetically barcoded antibodies
Abstract
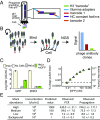
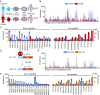
Human cells express thousands of different surface proteins that can be used for cell classification, or to distinguish healthy and disease conditions. A method capable of profiling a substantial fraction of the surface proteome simultaneously and inexpensively would enable more accurate and complete classification of cell states. We present a highly multiplexed and quantitative surface proteomic method using genetically barcoded antibodies called phage-antibody next-generation sequencing (PhaNGS). Using 144 preselected antibodies displayed on filamentous phage (Fab-phage) against 44 receptor targets, we assess changes in B cell surface proteins after the development of drug resistance in a patient with acute lymphoblastic leukemia (ALL) and in adaptation to oncogene expression in a Myc-inducible Burkitt lymphoma model. We further show PhaNGS can be applied at the single-cell level. Our results reveal that a common set of proteins including FLT3, NCR3LG1, and ROR1 dominate the response to similar oncogenic perturbations in B cells. Linking high-affinity, selective, genetically encoded binders to NGS enables direct and highly multiplexed protein detection, comparable to RNA-sequencing for mRNA. PhaNGS has the potential to profile a substantial fraction of the surface proteome simultaneously and inexpensively to enable more accurate and complete classification of cell states.
Keywords: NGS; biomarkers; cell surface proteomics; leukemia; phage display.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

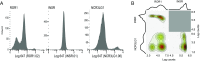
References
-
- Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. The technology and biology of single-cell RNA sequencing. Mol Cell. 2015;58:610–620. - PubMed
-
- Fan HC, Fu GK, Fodor SP. Expression profiling. Combinatorial labeling of single cells for gene expression cytometry. Science. 2015;347:1258367. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

