Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis
- PMID: 29476097
- PMCID: PMC5824882
- DOI: 10.1038/s41467-018-02892-y
Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis
Abstract
Fibroblasts regulate tissue homeostasis, coordinate inflammatory responses, and mediate tissue damage. In rheumatoid arthritis (RA), synovial fibroblasts maintain chronic inflammation which leads to joint destruction. Little is known about fibroblast heterogeneity or if aberrations in fibroblast subsets relate to pathology. Here, we show functional and transcriptional differences between fibroblast subsets from human synovial tissues using bulk transcriptomics of targeted subpopulations and single-cell transcriptomics. We identify seven fibroblast subsets with distinct surface protein phenotypes, and collapse them into three subsets by integrating transcriptomic data. One fibroblast subset, characterized by the expression of proteins podoplanin, THY1 membrane glycoprotein and cadherin-11, but lacking CD34, is threefold expanded in patients with RA relative to patients with osteoarthritis. These fibroblasts localize to the perivascular zone in inflamed synovium, secrete proinflammatory cytokines, are proliferative, and have an in vitro phenotype characteristic of invasive cells. Our strategy may be used as a template to identify pathogenic stromal cellular subsets in other complex diseases.
Conflict of interest statement
M.B.B., S.R., and C.D.B. declare that they received research funding from Roche, and M.B.B. is a consultant to Roche. The remaining authors declare no competing financial interests.
Figures
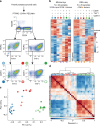
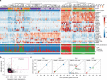


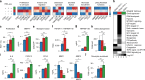
Comment in
-
Rheumatoid arthritis: RA synovium harbours distinct fibroblast subsets.Nat Rev Rheumatol. 2018 Apr 20;14(5):250. doi: 10.1038/nrrheum.2018.47. Nat Rev Rheumatol. 2018. PMID: 29674614 No abstract available.
-
A pathogenic hierarchy for synovial fibroblasts in rheumatoid arthritis.Ann Transl Med. 2018 Nov;6(Suppl 1):S75. doi: 10.21037/atm.2018.10.49. Ann Transl Med. 2018. PMID: 30613650 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 AR046713/AR/NIAMS NIH HHS/United States
- R01 CA170653/CA/NCI NIH HHS/United States
- P30 AR070253/AR/NIAMS NIH HHS/United States
- U19 AI111224/AI/NIAID NIH HHS/United States
- T32 HG002295/HG/NHGRI NIH HHS/United States
- R01 AR065538/AR/NIAMS NIH HHS/United States
- U01 GM092691/GM/NIGMS NIH HHS/United States
- F31 AR070582/AR/NIAMS NIH HHS/United States
- R01 AR063759/AR/NIAMS NIH HHS/United States
- U01 HG009379/HG/NHGRI NIH HHS/United States
- R01 AR063709/AR/NIAMS NIH HHS/United States
- Wellcome Trust/United Kingdom
- K08 AR063696/AR/NIAMS NIH HHS/United States
- UH2 AR067691/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

