Peatland Acidobacteria with a dissimilatory sulfur metabolism
- PMID: 29476143
- PMCID: PMC6018796
- DOI: 10.1038/s41396-018-0077-1
Peatland Acidobacteria with a dissimilatory sulfur metabolism
Abstract
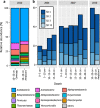
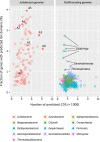
Sulfur-cycling microorganisms impact organic matter decomposition in wetlands and consequently greenhouse gas emissions from these globally relevant environments. However, their identities and physiological properties are largely unknown. By applying a functional metagenomics approach to an acidic peatland, we recovered draft genomes of seven novel Acidobacteria species with the potential for dissimilatory sulfite (dsrAB, dsrC, dsrD, dsrN, dsrT, dsrMKJOP) or sulfate respiration (sat, aprBA, qmoABC plus dsr genes). Surprisingly, the genomes also encoded DsrL, which so far was only found in sulfur-oxidizing microorganisms. Metatranscriptome analysis demonstrated expression of acidobacterial sulfur-metabolism genes in native peat soil and their upregulation in diverse anoxic microcosms. This indicated an active sulfate respiration pathway, which, however, might also operate in reverse for dissimilatory sulfur oxidation or disproportionation as proposed for the sulfur-oxidizing Desulfurivibrio alkaliphilus. Acidobacteria that only harbored genes for sulfite reduction additionally encoded enzymes that liberate sulfite from organosulfonates, which suggested organic sulfur compounds as complementary energy sources. Further metabolic potentials included polysaccharide hydrolysis and sugar utilization, aerobic respiration, several fermentative capabilities, and hydrogen oxidation. Our findings extend both, the known physiological and genetic properties of Acidobacteria and the known taxonomic diversity of microorganisms with a DsrAB-based sulfur metabolism, and highlight new fundamental niches for facultative anaerobic Acidobacteria in wetlands based on exploitation of inorganic and organic sulfur molecules for energy conservation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Meyer B, Imhoff JF, Kuever J. Molecular analysis of the distribution and phylogeny of the soxB gene among sulfur-oxidizing bacteria: evolution of the Sox sulfur oxidation enzyme system. Environ Microbiol. 2007;9:2957–77. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

