The evolutionary impact of intragenic FliA promoters in proteobacteria
- PMID: 29476659
- PMCID: PMC5943157
- DOI: 10.1111/mmi.13941
The evolutionary impact of intragenic FliA promoters in proteobacteria
Abstract
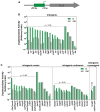
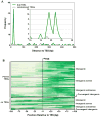
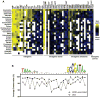
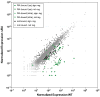
In Escherichia coli, one sigma factor recognizes the majority of promoters, and six 'alternative' sigma factors recognize specific subsets of promoters. The alternative sigma factor FliA (σ28 ) recognizes promoters upstream of many flagellar genes. We previously showed that most E. coli FliA binding sites are located inside genes. However, it was unclear whether these intragenic binding sites represent active promoters. Here, we construct and assay transcriptional promoter-lacZ fusions for all 52 putative FliA promoters previously identified by ChIP-seq. These experiments, coupled with integrative analysis of published genome-scale transcriptional datasets, strongly suggest that most intragenic FliA binding sites are active promoters that transcribe highly unstable RNAs. Additionally, we show that widespread intragenic FliA-dependent transcription may be a conserved phenomenon, but that specific promoters are not themselves conserved. We conclude that intragenic FliA-dependent promoters and the resulting RNAs are unlikely to have important regulatory functions. Nonetheless, one intragenic FliA promoter is broadly conserved and constrains evolution of the overlapping protein-coding gene. Thus, our data indicate that intragenic regulatory elements can influence bacterial protein evolution and suggest that the impact of intragenic regulatory sequences on genome evolution should be considered more broadly.
© 2018 John Wiley & Sons Ltd.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

