Epigenomic signature of adrenoleukodystrophy predicts compromised oligodendrocyte differentiation
- PMID: 29476661
- PMCID: PMC6857458
- DOI: 10.1111/bpa.12595
Epigenomic signature of adrenoleukodystrophy predicts compromised oligodendrocyte differentiation
Abstract
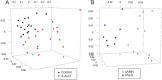
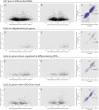
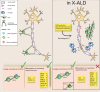
Epigenomic changes may either cause disease or modulate its expressivity, adding a layer of complexity to mendelian diseases. X-linked adrenoleukodystrophy (X-ALD) is a rare neurometabolic condition exhibiting discordant phenotypes, ranging from a childhood cerebral inflammatory demyelination (cALD) to an adult-onset mild axonopathy in spinal cords (AMN). The AMN form may occur with superimposed inflammatory brain demyelination (cAMN). All patients harbor loss of function mutations in the ABCD1 peroxisomal transporter of very-long chain fatty acids. The factors that account for the lack of genotype-phenotype correlation, even within the same family, remain largely unknown. To gain insight into this matter, here we compared the genome-wide DNA methylation profiles of morphologically intact frontal white matter areas of children affected by cALD with adult cAMN patients, including male controls in the same age group. We identified a common methylomic signature between the two phenotypes, comprising (i) hypermethylation of genes harboring the H3K27me3 mark at promoter regions, (ii) hypermethylation of genes with major roles in oligodendrocyte differentiation such as MBP, CNP, MOG and PLP1 and (iii) hypomethylation of immune-associated genes such as IFITM1 and CD59. Moreover, we found increased hypermethylation in CpGs of genes involved in oligodendrocyte differentiation, and also in genes with H3K27me3 marks in their promoter regions in cALD compared with cAMN, correlating with transcriptional and translational changes. Further, using a penalized logistic regression model, we identified the combined methylation levels of SPG20, UNC45A and COL9A3 and also, the combined expression levels of ID4 and MYRF to be good markers capable of discriminating childhood from adult inflammatory phenotypes. We thus propose the hypothesis that an epigenetically controlled, altered transcriptional program may drive an impaired oligodendrocyte differentiation and aberrant immune activation in X-ALD patients. These results shed light into disease pathomechanisms and uncover putative biomarkers of interest for prognosis and phenotypic stratification.
Keywords: adrenoleukodystrophy; epigenetics; myelin; neurodegeneration; oligodendrocytes; peroxisome.
© 2018 The Authors. Brain Pathology published by John Wiley & Sons Ltd on behalf of International Society of Neuropathology.
Conflict of interest statement
None declared
Figures






Similar articles
-
Functional genomic analysis unravels a metabolic-inflammatory interplay in adrenoleukodystrophy.Hum Mol Genet. 2012 Mar 1;21(5):1062-77. doi: 10.1093/hmg/ddr536. Epub 2011 Nov 17. Hum Mol Genet. 2012. PMID: 22095690 Free PMC article.
-
Abcd1 deficiency accelerates cuprizone-induced oligodendrocyte loss and axonopathy in a demyelinating mouse model of X-linked adrenoleukodystrophy.Acta Neuropathol Commun. 2023 Jun 18;11(1):98. doi: 10.1186/s40478-023-01595-w. Acta Neuropathol Commun. 2023. PMID: 37331971 Free PMC article.
-
Lack of adrenoleukodystrophy protein enhances oligodendrocyte disturbance and microglia activation in mice with combined Abcd1/Mag deficiency.Acta Neuropathol. 2007 Dec;114(6):573-86. doi: 10.1007/s00401-007-0288-4. Epub 2007 Sep 9. Acta Neuropathol. 2007. PMID: 17828604
-
Pathophysiology of X-linked adrenoleukodystrophy.Biochimie. 2014 Mar;98(100):135-42. doi: 10.1016/j.biochi.2013.11.023. Epub 2013 Dec 4. Biochimie. 2014. PMID: 24316281 Free PMC article. Review.
-
Management of adrenoleukodystrophy: From pre-clinical studies to the development of new therapies.Biomed Pharmacother. 2021 Nov;143:112214. doi: 10.1016/j.biopha.2021.112214. Epub 2021 Sep 21. Biomed Pharmacother. 2021. PMID: 34560537 Review.
Cited by
-
Deciphering the modifiers for phenotypic variability of X-linked adrenoleukodystrophy.World J Biol Chem. 2020 Nov 27;11(3):99-111. doi: 10.4331/wjbc.v11.i3.99. World J Biol Chem. 2020. PMID: 33274015 Free PMC article. Review.
-
Revisiting the Pathogenesis of X-Linked Adrenoleukodystrophy.Genes (Basel). 2025 May 17;16(5):590. doi: 10.3390/genes16050590. Genes (Basel). 2025. PMID: 40428412 Free PMC article. Review.
-
Controlled human exposures to diesel exhaust: a human epigenome-wide experiment of target bronchial epithelial cells.Environ Epigenet. 2021 Apr 9;7(1):dvab003. doi: 10.1093/eep/dvab003. eCollection 2021. Environ Epigenet. 2021. PMID: 33859829 Free PMC article.
-
The pathology of X-linked adrenoleukodystrophy: tissue specific changes as a clue to pathophysiology.Orphanet J Rare Dis. 2024 Mar 28;19(1):138. doi: 10.1186/s13023-024-03105-0. Orphanet J Rare Dis. 2024. PMID: 38549180 Free PMC article. Review.
-
Microglia damage precedes major myelin breakdown in X-linked adrenoleukodystrophy and metachromatic leukodystrophy.Glia. 2019 Jun;67(6):1196-1209. doi: 10.1002/glia.23598. Epub 2019 Feb 11. Glia. 2019. PMID: 30980503 Free PMC article.
References
-
- Anlar B, Waye JS, Eng B, Oguz KK (2006) Atypical clinical course in juvenile metachromatic leukodystrophy involving novel arylsulfatase A gene mutations. Dev Med Child Neurol 48:383–387. - PubMed
-
- Armada‐Moreira A, Ribeiro FF, Sebastião AM, Xapelli S (2015) Neuroinflammatory modulators of oligodendrogenesis. Neuroimmunol Neuroinflamm 2:263–273.
-
- Asheuer M, Bieche I, Laurendeau I, Moser A, Hainque B, Vidaud M, Aubourg P (2005) Decreased expression of ABCD4 and BG1 genes early in the pathogenesis of X‐linked adrenoleukodystrophy. Hum Mol Genet 14:1293–1303. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

