Dopamine D1 Receptor-Positive Neurons in the Lateral Nucleus of the Cerebellum Contribute to Cognitive Behavior
- PMID: 29478701
- PMCID: PMC6072628
- DOI: 10.1016/j.biopsych.2018.01.019
Dopamine D1 Receptor-Positive Neurons in the Lateral Nucleus of the Cerebellum Contribute to Cognitive Behavior
Abstract
Background: Studies in humans and nonhuman primates have identified a region of the dentate nucleus of the cerebellum, or the lateral cerebellar nucleus (LCN) in rodents, activated during performance of cognitive tasks involving complex spatial and sequential planning. Whether such a subdivision exists in rodents is not known. Dopamine and its receptors, which are implicated in cognitive function, are present in the cerebellar nuclei, but their function is unknown.
Methods: Using viral and genetic strategies in mice, we examined cellular phenotypes of dopamine D1 receptor-positive (D1R+) cells in the LCN with whole-cell patch clamp recordings, messenger RNA profiling, and immunohistochemistry to examine D1R expression in mouse LCN and human dentate nucleus of the cerebellum. We used chemogenetics to inhibit D1R+ neurons and examined behaviors including spatial navigation, social recognition memory, prepulse inhibition of the acoustic startle reflex, response inhibition, and working memory to test the necessity of these neurons in these behaviors.
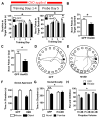
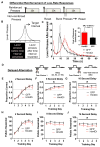
Results: We identified a population of D1R+ neurons that are localized to an anatomically distinct region of the LCN. We also observed D1R+ neurons in human dentate nucleus of the cerebellum, which suggests an evolutionarily conserved population of dopamine-receptive neurons in this region. The genetic, electrophysiological, and anatomical profile of mouse D1R neurons is consistent with a heterogeneous population of gamma-aminobutyric acidergic, and to a lesser extent glutamatergic, cell types. Selective inhibition of D1R+ LCN neurons impairs spatial navigation memory, response inhibition, working memory, and prepulse inhibition of the acoustic startle reflex.
Conclusions: Collectively, these data demonstrate a functional link between genetically distinct neurons in the LCN and cognitive behaviors.
Keywords: Cerebellar nuclei; Cerebellum; Cognition; DREADD receptor; Dopamine D(1) receptor; RiboTag.
Copyright © 2018 Society of Biological Psychiatry. Published by Elsevier Inc. All rights reserved.
Figures

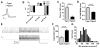


References
-
- Kawato M, Gomi H. A computational model of four regions of the cerebellum based on feedback-error learning. Biological cybernetics. 1992;68:95–103. - PubMed
-
- Medina JF, Nores WL, Ohyama T, Mauk MD. Mechanisms of cerebellar learning suggested by eyelid conditioning. Current opinion in neurobiology. 2000;10:717–724. - PubMed
-
- Braak E, Arai K, Braak H. Cerebellar involvement in Pick’s disease: affliction of mossy fibers, monodendritic brush cells, and dentate projection neurons. Experimental neurology. 1999;159:153–163. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

