Dampened STING-Dependent Interferon Activation in Bats
- PMID: 29478775
- PMCID: PMC7104992
- DOI: 10.1016/j.chom.2018.01.006
Dampened STING-Dependent Interferon Activation in Bats
Abstract
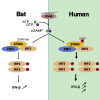
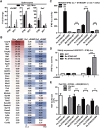
Compared with terrestrial mammals, bats have a longer lifespan and greater capacity to co-exist with a variety of viruses. In addition to cytosolic DNA generated by these viral infections, the metabolic demands of flight cause DNA damage and the release of self-DNA into the cytoplasm. However, whether bats have an altered DNA sensing/defense system to balance high cytosolic DNA levels remains an open question. We demonstrate that bats have a dampened interferon response due to the replacement of the highly conserved serine residue (S358) in STING, an essential adaptor protein in multiple DNA sensing pathways. Reversing this mutation by introducing S358 restored STING functionality, resulting in interferon activation and virus inhibition. Combined with previous reports on bat-specific changes of other DNA sensors such as TLR9, IFI16, and AIM2, our findings shed light on bat adaptation to flight, their long lifespan, and their unique capacity to serve as a virus reservoir.
Keywords: DNA sensing; STING; bats; dampened; interferon; virus.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures

References
-
- Barzilai A., Rotman G., Shiloh Y. ATM deficiency and oxidative stress: a new dimension of defective response to DNA damage. DNA Repair. 2002;1:3–25. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

