Genome-wide CRISPR-Cas9 Screen Identifies Leukemia-Specific Dependence on a Pre-mRNA Metabolic Pathway Regulated by DCPS
- PMID: 29478914
- PMCID: PMC5849534
- DOI: 10.1016/j.ccell.2018.01.012
Genome-wide CRISPR-Cas9 Screen Identifies Leukemia-Specific Dependence on a Pre-mRNA Metabolic Pathway Regulated by DCPS
Abstract
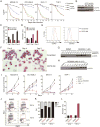
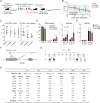
To identify novel targets for acute myeloid leukemia (AML) therapy, we performed genome-wide CRISPR-Cas9 screening using AML cell lines, followed by a second screen in vivo. Here, we show that the mRNA decapping enzyme scavenger (DCPS) gene is essential for AML cell survival. The DCPS enzyme interacted with components of pre-mRNA metabolic pathways, including spliceosomes, as revealed by mass spectrometry. RG3039, a DCPS inhibitor originally developed to treat spinal muscular atrophy, exhibited anti-leukemic activity via inducing pre-mRNA mis-splicing. Humans harboring germline biallelic DCPS loss-of-function mutations do not exhibit aberrant hematologic phenotypes, indicating that DCPS is dispensable for human hematopoiesis. Our findings shed light on a pre-mRNA metabolic pathway and identify DCPS as a target for AML therapy.
Keywords: CRISPR-Cas9 saturation mutagenesis; acute myeloid leukemia; decapping enzyme; drug repurposing; genome-wide CRISPR-Cas9 screening; mRNA decay; pre-mRNA metabolism; pre-mRNA splicing.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures




Comment in
-
Targeting mRNA Decapping in AML.Cancer Cell. 2018 Mar 12;33(3):339-341. doi: 10.1016/j.ccell.2018.02.015. Cancer Cell. 2018. PMID: 29533777 Free PMC article.
References
-
- Abifadel M, Varret M, Rabès J-P, Allard D, Ouguerram K, Devillers M, Cruaud C, Benjannet S, Wickham L, Erlich D, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–156. - PubMed
-
- Boehm JS, Hahn WC. Towards systematic functional characterization of cancer genomes. Nat Rev Genet. 2011;12:487–498. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

