Plant Materials are Sustainable Substrates Supporting New Technologies of Plant-Only-Based Culture Media for in vitro Culturing of the Plant Microbiota
- PMID: 29479006
- PMCID: PMC5877342
- DOI: 10.1264/jsme2.ME17135
Plant Materials are Sustainable Substrates Supporting New Technologies of Plant-Only-Based Culture Media for in vitro Culturing of the Plant Microbiota
Abstract
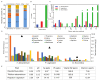
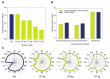
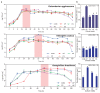
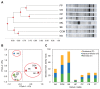
In order to improve the culturability and biomass production of rhizobacteria, we previously introduced plant-only-based culture media. We herein attempted to widen the scope of plant materials suitable for the preparation of plant-only-based culture media. We chemically analyzed the refuse of turfgrass, cactus, and clover. They were sufficiently rich to support good in vitro growth by rhizobacteria isolates representing Proteobacteria and Firmicutes. They were also adequate and efficient to produce a cell biomass in liquid batch cultures. These culture media were as sufficient as artificial culture media for the cultivation and recovery of the in situ rhizobacteria of barley (Hordeum murinum L.). Based on culture-dependent (CFU plate counting) and culture-independent analyses (qPCR), mowed turfgrass, in particular, supported the highest culturable population of barley endophytes, representing >16% of the total bacterial number quantified with qPCR. This accurately reflected the endophytic community composition, in terms of diversity indices (S', H', and D') based on PCR-DGGE, and clustered the plant culture media together with the qPCR root populations away from the artificial culture media. Despite the promiscuous nature of the plant materials tested to culture the plant microbiome, our results indicated that plant materials of a homologous nature to the tested host plant, at least at the family level, and/or of the same environment were more likely to be selected. Plant-only-based culture media require further refinements in order to provide selectivity for the in vitro growth of members of the plant microbiome, particularly difficult-to-culture bacteria. This will provide insights into their hidden roles in the environment and support future culturomic studies.
Keywords: microbial biomass; plant microbiome/microbiota; plant-only-based culture media; rhizobacteria culturability; wild desert barley.
Figures
References
-
- Abdel-Rahman M.A., Tashiro Y., Sonomoto K. Recent advances in lactic acid production by microbial fermentation processes. Biotechnol Adv. 2013;31:877–902. - PubMed
-
- Ali S.M., Amin G., Fayez M., El-Tahan M., Monib M., Hegazi N.A. Production of rhizobia biofertilizers using baker’s yeast effluent and their application to Leucaena leucocephala. Arch Agron Soil Sci. 2005;51:605–617.
-
- Ali S.M., Hamza M.A., Amin G., Fayez M., El-Tahan M., Monib M., Hegazi N.A. Production of biofertilizers using baker’s yeast effluent and their application to wheat and barley grown in north Sinai deserts. Arch Agron Soil Sci. 2005;51:589–604.
-
- Austin B. The value of cultures to modern microbiology. In: Hedlund B.P., Sutcliffe I.C., Trujillo M.E., editors. Antonie Van Leeuwenhoek. Vol. 110. Springer; New York: 2017. pp. 1247–1256. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

