Integrative omics for health and disease
- PMID: 29479082
- PMCID: PMC5990367
- DOI: 10.1038/nrg.2018.4
Integrative omics for health and disease
Abstract
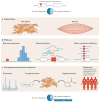
Advances in omics technologies - such as genomics, transcriptomics, proteomics and metabolomics - have begun to enable personalized medicine at an extraordinarily detailed molecular level. Individually, these technologies have contributed medical advances that have begun to enter clinical practice. However, each technology individually cannot capture the entire biological complexity of most human diseases. Integration of multiple technologies has emerged as an approach to provide a more comprehensive view of biology and disease. In this Review, we discuss the potential for combining diverse types of data and the utility of this approach in human health and disease. We provide examples of data integration to understand, diagnose and inform treatment of diseases, including rare and common diseases as well as cancer and transplant biology. Finally, we discuss technical and other challenges to clinical implementation of integrative omics.
Conflict of interest statement
The authors declare competing interests. See Web version for details.
Figures


References
-
- Worthey EA, et al. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011;13:255–262. This is the first paper describing the treatment-changing diagnosis in an individual patient using exome sequencing, paving the way for clinical applications of genomics. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

