Serine is the major residue for ADP-ribosylation upon DNA damage
- PMID: 29480802
- PMCID: PMC5837557
- DOI: 10.7554/eLife.34334
Serine is the major residue for ADP-ribosylation upon DNA damage
Abstract
Poly(ADP-ribose) polymerases (PARPs) are a family of enzymes that synthesise ADP-ribosylation (ADPr), a reversible modification of proteins that regulates many different cellular processes. Several mammalian PARPs are known to regulate the DNA damage response, but it is not clear which amino acids in proteins are the primary ADPr targets. Previously, we reported that ARH3 reverses the newly discovered type of ADPr (ADPr on serine residues; Ser-ADPr) and developed tools to analyse this modification (Fontana et al., 2017). Here, we show that Ser-ADPr represents the major fraction of ADPr synthesised after DNA damage in mammalian cells and that globally Ser-ADPr is dependent on HPF1, PARP1 and ARH3. In the absence of HPF1, glutamate/aspartate becomes the main target residues for ADPr. Furthermore, we describe a method for site-specific validation of serine ADP-ribosylated substrates in cells. Our study establishes serine as the primary form of ADPr in DNA damage signalling.
Keywords: ADP-ribosylation; ARH3; DNA damage; DNA repair; PARP; biochemistry; chemical biology; enzyme; human.
© 2018, Palazzo et al.
Conflict of interest statement
LP, OL, EP, HD, IM, IA No competing interests declared
Figures



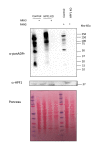
References
-
- Bilan V, Leutert M, Nanni P, Panse C, Hottiger MO. Combining higher-energy collision dissociation and electron-transfer/higher-energy collision dissociation fragmentation in a product-dependent manner confidently assigns proteomewide ADP-ribose acceptor sites. Analytical Chemistry. 2017;89:1523–1530. doi: 10.1021/acs.analchem.6b03365. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

