Integrated RNA and DNA sequencing reveals early drivers of metastatic breast cancer
- PMID: 29480819
- PMCID: PMC5873890
- DOI: 10.1172/JCI96153
Integrated RNA and DNA sequencing reveals early drivers of metastatic breast cancer
Abstract
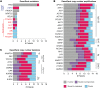
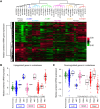
Breast cancer metastasis remains a clinical challenge, even within a single patient across multiple sites of the disease. Genome-wide comparisons of both the DNA and gene expression of primary tumors and metastases in multiple patients could help elucidate the underlying mechanisms that cause breast cancer metastasis. To address this issue, we performed DNA exome and RNA sequencing of matched primary tumors and multiple metastases from 16 patients, totaling 83 distinct specimens. We identified tumor-specific drivers by integrating known protein-protein network information with RNA expression and somatic DNA alterations and found that genetic drivers were predominantly established in the primary tumor and maintained through metastatic spreading. In addition, our analyses revealed that most genetic drivers were DNA copy number changes, the TP53 mutation was a recurrent founding mutation regardless of subtype, and that multiclonal seeding of metastases was frequent and occurred in multiple subtypes. Genetic drivers unique to metastasis were identified as somatic mutations in the estrogen and androgen receptor genes. These results highlight the complexity of metastatic spreading, be it monoclonal or multiclonal, and suggest that most metastatic drivers are established in the primary tumor, despite the substantial heterogeneity seen in the metastases.
Keywords: Breast cancer; Oncology.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

